Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression
- PMID: 28680090
- PMCID: PMC5498532
- DOI: 10.1038/s41598-017-04234-2
Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression
Abstract
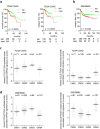
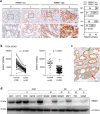
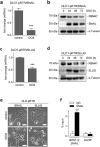
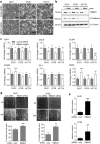
Epithelial-mesenchymal transition (EMT) plays an important role in tumor invasion and metastasis. A comprehensive, bioinformatics analysis of CCLE and TCGA datasets of seven tumor types allowed us to identify a novel pan-cancer EMT-associated gene expression signature consisting of 16 epithelial and 4 mesenchymal state-associated mRNAs. Among the identified epithelial cell state-associated factors, down-regulation of the RBM47 (RNA binding motif protein 47) mRNA displayed the most significant association with metastasis and poor survival in multiple cohorts of colorectal cancer (CRC) patients. Moreover, decreased RBM47 protein expression was associated with metastasis in a cohort of primary CRCs. RBM47 was directly suppressed during EMT induced by IL6-activated STAT3 or ectopic SNAIL and SLUG expression via conserved binding motifs of these factors within the RBM47 promoter. Moreover, RNAi-mediated down-regulation of RBM47 in CRC lines resulted in increased cell migration, invasion and metastases formation. As demonstrated by the example of RBM47, the EMT-associated signature characterized here allows to identify biomarkers for predicting clinical outcome of CRC and presumably other cancer entities. In addition, our functional analysis of RBM47 shows that the down-regulation of RBM47 during CRC progression may promote EMT and metastasis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Genetic and epigenetic down-regulation of microRNA-212 promotes colorectal tumor metastasis via dysregulation of MnSOD.Gastroenterology. 2013 Aug;145(2):426-36.e1-6. doi: 10.1053/j.gastro.2013.04.004. Epub 2013 Apr 9. Gastroenterology. 2013. PMID: 23583431
-
Copy number amplification-induced overexpression of lncRNA LOC101927668 facilitates colorectal cancer progression by recruiting hnRNPD to disrupt RBM47/p53/p21 signaling.J Exp Clin Cancer Res. 2024 Sep 30;43(1):274. doi: 10.1186/s13046-024-03193-7. J Exp Clin Cancer Res. 2024. PMID: 39350250 Free PMC article.
-
Lipocalin2 suppresses metastasis of colorectal cancer by attenuating NF-κB-dependent activation of snail and epithelial mesenchymal transition.Mol Cancer. 2016 Dec 3;15(1):77. doi: 10.1186/s12943-016-0564-9. Mol Cancer. 2016. PMID: 27912767 Free PMC article.
-
Pharmacological Targeting of Epithelial-to-Mesenchymal Transition in Colorectal Cancer.Curr Pharm Des. 2022;28(28):2298-2311. doi: 10.2174/1381612828666220728152350. Curr Pharm Des. 2022. PMID: 35909286 Review.
-
Current Progress of EMT: A New Direction of Targeted Therapy for Colorectal Cancer with Invasion and Metastasis.Biomolecules. 2022 Nov 22;12(12):1723. doi: 10.3390/biom12121723. Biomolecules. 2022. PMID: 36551152 Free PMC article. Review.
Cited by
-
Rapid proteomic analysis for solid tumors reveals LSD1 as a drug target in an end-stage cancer patient.Mol Oncol. 2018 Aug;12(8):1296-1307. doi: 10.1002/1878-0261.12326. Epub 2018 Jun 14. Mol Oncol. 2018. PMID: 29901861 Free PMC article.
-
RNA binding motif 47 (RBM47): emerging roles in vertebrate development, RNA editing and cancer.Mol Cell Biochem. 2021 Dec;476(12):4493-4505. doi: 10.1007/s11010-021-04256-5. Epub 2021 Sep 9. Mol Cell Biochem. 2021. PMID: 34499322 Review.
-
The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity.NAR Cancer. 2020 Dec;2(4):zcaa027. doi: 10.1093/narcan/zcaa027. Epub 2020 Oct 9. NAR Cancer. 2020. PMID: 33094286 Free PMC article.
-
Construction of a Pearson- and MIC-Based Co-expression Network to Identify Potential Cancer Genes.Interdiscip Sci. 2022 Mar;14(1):245-257. doi: 10.1007/s12539-021-00485-w. Epub 2021 Oct 25. Interdiscip Sci. 2022. PMID: 34694561
-
An Unsupervised Strategy for Identifying Epithelial-Mesenchymal Transition State Metrics in Breast Cancer and Melanoma.iScience. 2020 May 22;23(5):101080. doi: 10.1016/j.isci.2020.101080. Epub 2020 Apr 22. iScience. 2020. PMID: 32371374 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

