Molecular Characterization of MYB28 Involved in Aliphatic Glucosinolate Biosynthesis in Chinese Kale (Brassica oleracea var. alboglabra Bailey)
- PMID: 28680435
- PMCID: PMC5478679
- DOI: 10.3389/fpls.2017.01083
Molecular Characterization of MYB28 Involved in Aliphatic Glucosinolate Biosynthesis in Chinese Kale (Brassica oleracea var. alboglabra Bailey)
Abstract
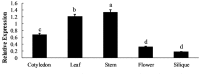
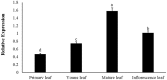
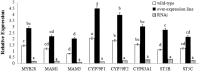
Glucosinolates are Brassicaceae-specific secondary metabolites that act as crop protectants, flavor precursors, and cancer-prevention agents, which shows strong evidences of anticarcinogentic, antioxidant, and antimicrobial activities. MYB28, the R2R3-MYB28 transcription factor, directly activates genes involved in aliphatic glucosinolate biosynthesis. In this study, the MYB28 homology (BoaMYB28) was identified in Chinese kale (Brassica oleracea var. alboglabra Bailey). Analysis of the nucleotide sequence indicated that the cDNA of BoaMYB28 was 1257 bp with an ORF of 1020 bp. The deduced BoaMYB28 protein was a polypeptide of 339 amino acid with a putative molecular mass of 38 kDa and a pI of 6.87. Sequence homology and phylogenetic analysis showed that BoaMYB28 was most closely related to MYB28 homologs from the Brassicaceae family. The expression levels of BoaMYB28 varies across the tissues and developmental stages. BoaMYB28 transcript levels were higher in leaves and stems compared with those in cotyledons, flowers, and siliques. BoaMYB28 was expressed across all developmental leaf stages, with higher transcript accumulation in mature and inflorescence leaves. Over-expression and RNAi studies showed that BoaMYB28 retains the basic MYB28 gene function as a major transcriptional regulator of aliphatic glucosinolate pathway. The results indicated that over-expression and RNAi lines showed no visible difference on plant morphology. The contents of aliphatic glucosinolates and transcript levels of aliphatic glucosinolate biosynthesis genes increased in over-expression lines and decreased in RNAi lines. In over-expression lines, aliphatic glucosinolate contents were 1.5- to 3-fold higher than those in the wild-type, while expression levels of aliphatic glucosinolate biosynthesis genes were 1.5- to 4-fold higher than those in the wild-type. In contrast, the contents of aliphatic glucosinolates and transcript levels of aliphatic glucosinolate biosynthesis genes in RNAi lines were considerably lower than those in the wild-type. The results suggest that BoaMYB28 has the potential to alter the aliphatic glucosinolates contents in Chinese kale at the genetic level.
Keywords: Chinese kale; RNAi; aliphatic glucosinolate; over-expression; transcription factors.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

