Logic Modeling in Quantitative Systems Pharmacology
- PMID: 28681552
- PMCID: PMC5572374
- DOI: 10.1002/psp4.12225
Logic Modeling in Quantitative Systems Pharmacology
Abstract
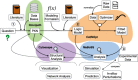
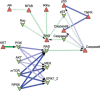
Here we present logic modeling as an approach to understand deregulation of signal transduction in disease and to characterize a drug's mode of action. We discuss how to build a logic model from the literature and experimental data and how to analyze the resulting model to obtain insights of relevance for systems pharmacology. Our workflow uses the free tools OmniPath (network reconstruction from the literature), CellNOpt (model fit to experimental data), MaBoSS (model analysis), and Cytoscape (visualization).
© 2017 The Authors CPT: Pharmacometrics & Systems Pharmacology published by Wiley Periodicals, Inc. on behalf of American Society for Clinical Pharmacology and Therapeutics.
Figures




Similar articles
-
Construction of cell type-specific logic models of signaling networks using CellNOpt.Methods Mol Biol. 2013;930:179-214. doi: 10.1007/978-1-62703-059-5_8. Methods Mol Biol. 2013. PMID: 23086842
-
Use of mathematics to guide target selection in systems pharmacology; application to receptor tyrosine kinase (RTK) pathways.Eur J Pharm Sci. 2017 Nov 15;109S:S140-S148. doi: 10.1016/j.ejps.2017.05.049. Epub 2017 May 24. Eur J Pharm Sci. 2017. PMID: 28549678
-
Boolean Modeling in Quantitative Systems Pharmacology: Challenges and Opportunities.Crit Rev Biomed Eng. 2019;47(6):473-488. doi: 10.1615/CritRevBiomedEng.2020030796. Crit Rev Biomed Eng. 2019. PMID: 32421972 Review.
-
Drug effects viewed from a signal transduction network perspective.J Med Chem. 2009 Dec 24;52(24):8038-46. doi: 10.1021/jm901001p. J Med Chem. 2009. PMID: 19891439
-
Merging systems biology with pharmacodynamics.Sci Transl Med. 2012 Mar 21;4(126):126ps7. doi: 10.1126/scitranslmed.3003563. Sci Transl Med. 2012. PMID: 22440734 Free PMC article. Review.
Cited by
-
Unveiling the signaling network of FLT3-ITD AML improves drug sensitivity prediction.Elife. 2024 Apr 2;12:RP90532. doi: 10.7554/eLife.90532. Elife. 2024. PMID: 38564252 Free PMC article.
-
Network-Based Systems Analysis Explains Sequence-Dependent Synergism of Bortezomib and Vorinostat in Multiple Myeloma.AAPS J. 2021 Aug 17;23(5):101. doi: 10.1208/s12248-021-00622-9. AAPS J. 2021. PMID: 34403034
-
LM-Merger: A workflow for merging logical models with an application to gene regulation.bioRxiv [Preprint]. 2024 Dec 17:2024.09.13.612961. doi: 10.1101/2024.09.13.612961. bioRxiv. 2024. Update in: BMC Bioinformatics. 2025 Jul 15;26(1):178. doi: 10.1186/s12859-025-06212-2. PMID: 39345612 Free PMC article. Updated. Preprint.
-
Boolean network modeling in systems pharmacology.J Pharmacokinet Pharmacodyn. 2018 Feb;45(1):159-180. doi: 10.1007/s10928-017-9567-4. Epub 2018 Jan 6. J Pharmacokinet Pharmacodyn. 2018. PMID: 29307099 Free PMC article. Review.
-
Gegen Qinlian decoction enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by remodelling the gut microbiota and the tumour microenvironment.Cell Death Dis. 2019 May 28;10(6):415. doi: 10.1038/s41419-019-1638-6. Cell Death Dis. 2019. PMID: 31138779 Free PMC article.
References
-
- Swinney, D.C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519 (2011). - PubMed
-
- Iorns, E. , Lord, C.J. , Turner, N. & Ashworth, A. Utilizing RNA interference to enhance cancer drug discovery. Nat. Rev. Drug Discov. 6, 556–568 (2007). - PubMed
-
- Shembekar, N. , Chaipan, C. , Utharala, R. & Merten, C.A. Droplet‐based microfluidics in drug discovery, transcriptomics and high‐throughput molecular genetics. Lab Chip 16, 1314–1331 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

