CRY1/2 Selectively Repress PPARδ and Limit Exercise Capacity
- PMID: 28683290
- PMCID: PMC5546250
- DOI: 10.1016/j.cmet.2017.06.002
CRY1/2 Selectively Repress PPARδ and Limit Exercise Capacity
Abstract
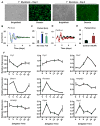
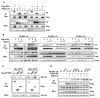
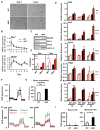
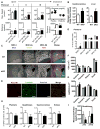
Cellular metabolite balance and mitochondrial function are under circadian control, but the pathways connecting the molecular clock to these functions are unclear. Peroxisome proliferator-activated receptor delta (PPARδ) enables preferential utilization of lipids as fuel during exercise and is a major driver of exercise endurance. We show here that the circadian repressors CRY1 and CRY2 function as co-repressors for PPARδ. Cry1-/-;Cry2-/- myotubes and muscles exhibit elevated expression of PPARδ target genes, particularly in the context of exercise. Notably, CRY1/2 seem to repress a distinct subset of PPARδ target genes in muscle compared to the co-repressor NCOR1. In vivo, genetic disruption of Cry1 and Cry2 enhances sprint exercise performance in mice. Collectively, our data demonstrate that CRY1 and CRY2 modulate exercise physiology by altering the activity of several transcription factors, including CLOCK/BMAL1 and PPARδ, and thereby alter energy storage and substrate selection for energy production.
Keywords: CRY1; CRY2; PPAR; beta oxidation; circadian; clock; cryptochrome; exercise; muscle; sprint.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
References
-
- Abe T, Kitaoka Y, Kikuchi DM, Takeda K, Numata O, Takemasa T. High-intensity interval training-induced metabolic adaptation coupled with an increase in Hif-1alpha and glycolytic protein expression. Journal of applied physiology. 2015;119:1297–1302. - PubMed
-
- Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G. Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1alpha. Cell metabolism. 2017;25:93–101. - PubMed
-
- Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19090–19095. - PMC - PubMed
-
- Bae K, Lee K, Seo Y, Lee H, Kim D, Choi I. Differential effects of two period genes on the physiology and proteomic profiles of mouse anterior tibialis muscles. Molecules and cells. 2006;22:275–284. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

