Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors
- PMID: 28683294
- PMCID: PMC5555084
- DOI: 10.1016/j.cmet.2017.06.004
Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors
Abstract
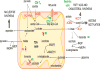
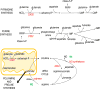
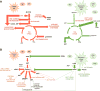
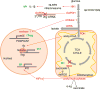
It has been appreciated for nearly 100 years that cancer cells are metabolically distinct from resting tissues. More recently understood is that this metabolic phenotype is not unique to cancer cells but instead reflects characteristics of proliferating cells. Similar metabolic transitions also occur in the immune system as cells transition from resting state to stimulated effectors. A key finding in immune metabolism is that the metabolic programs of different cell subsets are distinctly associated with immunological function. Further, interruption of those metabolic pathways can shift immune cell fate to modulate immunity. These studies have identified numerous metabolic similarities between cancer and immune cells but also critical differences that may be exploited and that affect treatment of cancer and immunological diseases.
Keywords: T cell; antitumor immunity; glycolysis; inflammation; macrophage; mitochondria; tumor metabolism; tumor microenvironment.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

References
-
- Abolmaali SS, Tamaddon AM, Dinarvand R. A review of therapeutic challenges and achievements of methotrexate delivery systems for treatment of cancer and rheumatoid arthritis. Cancer Chemother. Pharmacol. 2013;71:1115–1130. - PubMed
-
- Acuto O, Michel F. CD28-mediated co-stimulation: a quantitative support for TCR signalling. Nat Rev Immunol. 2003;3:939–951. - PubMed
-
- Allison AC. Immunosuppressive drugs: the first 50 years and a glance forward. Immunopharmacology. 2000;47:63–83. - PubMed
-
- Appleman LJ, van Puijenbroek AAFL, Shu KM, Nadler LM, Boussiotis VA. CD28 Costimulation Mediates Down-Regulation of p27kip1 and Cell Cycle Progression by Activation of the PI3K/PKB Signaling Pathway in Primary Human T Cells. J. Immunol. 2002;168:2729–2736. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

