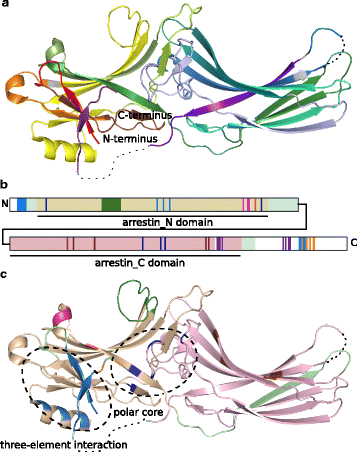
Uncovering missing pieces: duplication and deletion history of arrestins in deuterostomes
- PMID: 28683816
- PMCID: PMC5501109
- DOI: 10.1186/s12862-017-1001-4
Uncovering missing pieces: duplication and deletion history of arrestins in deuterostomes
Abstract
Background: The cytosolic arrestin proteins mediate desensitization of activated G protein-coupled receptors (GPCRs) via competition with G proteins for the active phosphorylated receptors. Arrestins in active, including receptor-bound, conformation are also transducers of signaling. Therefore, this protein family is an attractive therapeutic target. The signaling outcome is believed to be a result of structural and sequence-dependent interactions of arrestins with GPCRs and other protein partners. Here we elucidated the detailed evolution of arrestins in deuterostomes.
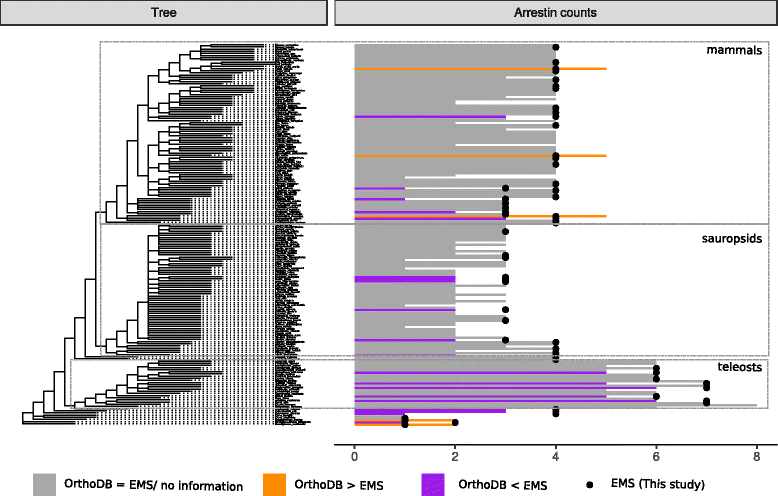
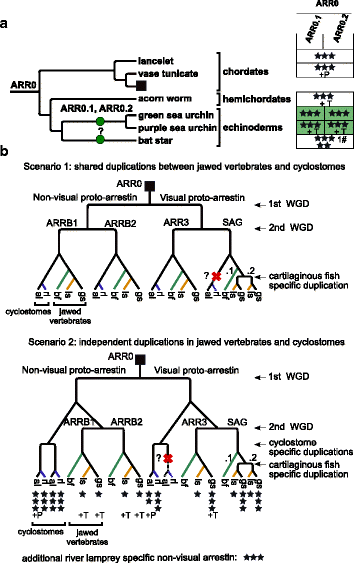
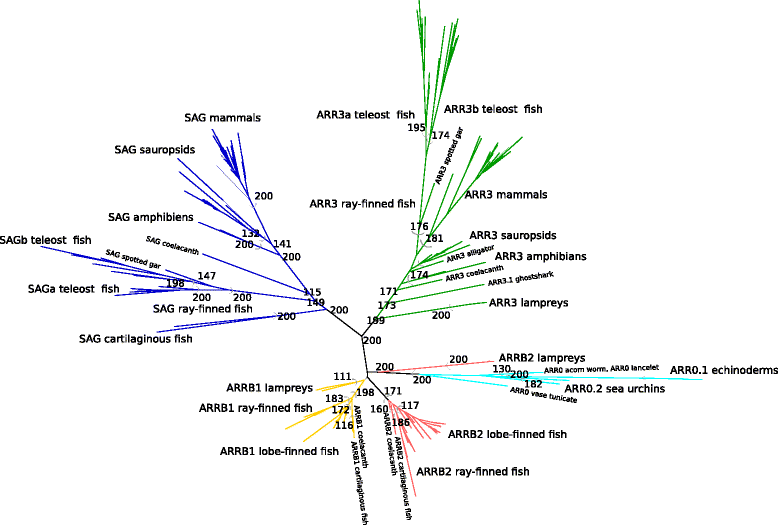
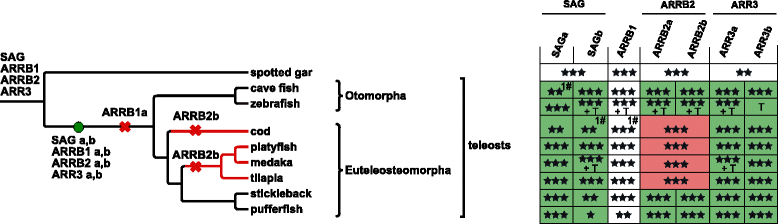
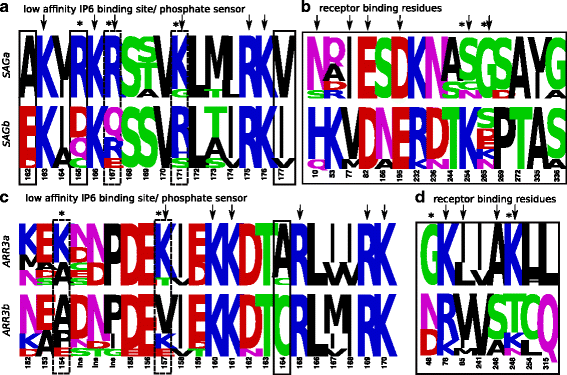
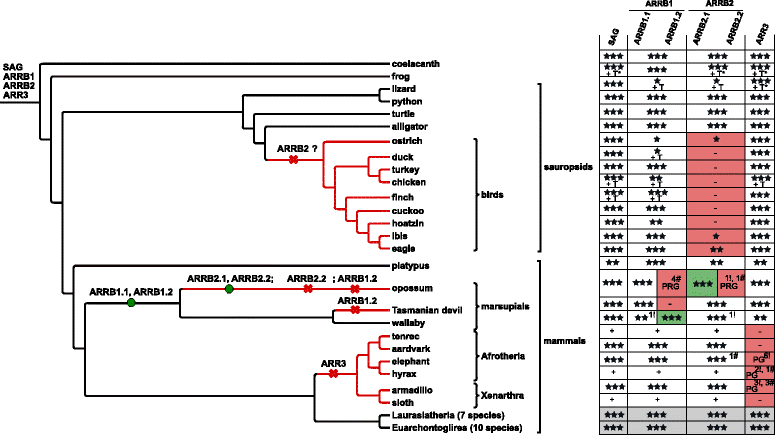
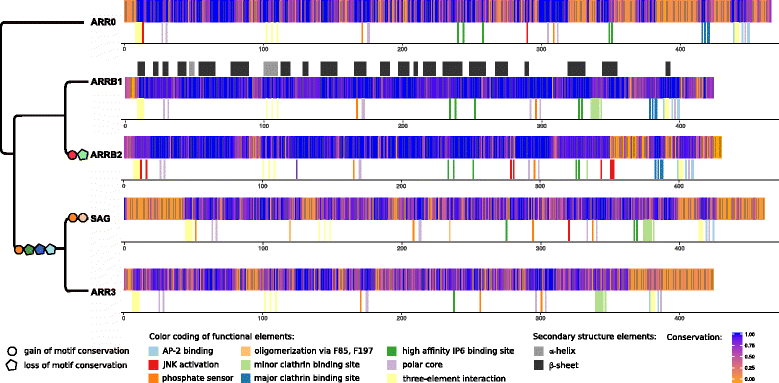
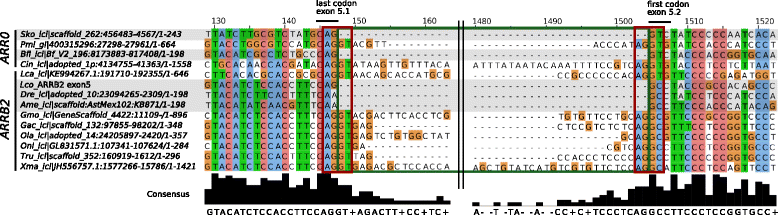
Results: Identity and number of arrestin paralogs were determined searching deuterostome genomes and gene expression data. In contrast to standard gene prediction methods, our strategy first detects exons situated on different scaffolds and then solves the problem of assigning them to the correct gene. This increases both the completeness and the accuracy of the annotation in comparison to conventional database search strategies applied by the community. The employed strategy enabled us to map in detail the duplication- and deletion history of arrestin paralogs including tandem duplications, pseudogenizations and the formation of retrogenes. The two rounds of whole genome duplications in the vertebrate stem lineage gave rise to four arrestin paralogs. Surprisingly, visual arrestin ARR3 was lost in the mammalian clades Afrotheria and Xenarthra. Duplications in specific clades, on the other hand, must have given rise to new paralogs that show signatures of diversification in functional elements important for receptor binding and phosphate sensing.
Conclusion: The current study traces the functional evolution of deuterostome arrestins in unprecedented detail. Based on a precise re-annotation of the exon-intron structure at nucleotide resolution, we infer the gain and loss of paralogs and patterns of conservation, co-variation and selection.
Keywords: Arrestin; Evolution; Gene duplication; Receptor specificity; Retrogene; Signaling.
Figures


References
-
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, Xu Q, Waal PWd, Ke J, Tan MHE, Zhang C, Moeller A, West GM, Pascal BD, van Eps N, Caro LN, Vishnivetskiy SA, Lee RJ, Suino-Powell KM, Gu X, Pal K, Ma J, Zhi X, Boutet S, Williams GJ, Messerschmidt M, Gati C, Zatsepin NA, Wang D, James D, Basu S, Roy-Chowdhury S, Conrad CE, Coe J, Liu H, Lisova S, Kupitz C, Grotjohann I, Fromme R, Jiang Y, Tan M, Yang H, Li J, Wang M, Zheng Z, Li D, Howe N, Zhao Y, Standfuss J, Diederichs K, Dong Y, Potter CS, Carragher B, Caffrey M, Jiang H, Chapman HN, Spence JCH, Fromme P, Weierstall U, Ernst OP, Katritch V, Gurevich VV, Griffin PR, Hubbell WL, Stevens RC, Cherezov V, Melcher K, Xu HE. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523(7562):561–7. doi: 10.1038/nature14656. - DOI - PMC - PubMed
-
- Luttrell LM. Molecular Biology of Arrestins. Progress in molecular biology and translational science, vol. v. 118. Oxford: Elsevier Science & Technology; 2013. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

