Dynamic ubiquitin signaling in cell cycle regulation
- PMID: 28684425
- PMCID: PMC5551716
- DOI: 10.1083/jcb.201703170
Dynamic ubiquitin signaling in cell cycle regulation
Abstract
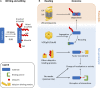
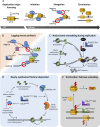
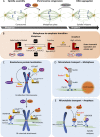
The cell division cycle is driven by a collection of enzymes that coordinate DNA duplication and separation, ensuring that genomic information is faithfully and perpetually maintained. The activity of the effector proteins that perform and coordinate these biological processes oscillates by regulated expression and/or posttranslational modifications. Ubiquitylation is a cardinal cellular modification and is long known for driving cell cycle transitions. In this review, we emphasize emerging concepts of how ubiquitylation brings the necessary dynamicity and plasticity that underlie the processes of DNA replication and mitosis. New studies, often focusing on the regulation of chromosomal proteins like DNA polymerases or kinetochore kinases, are demonstrating that ubiquitylation is a versatile modification that can be used to fine-tune these cell cycle events, frequently through processes that do not involve proteasomal degradation. Understanding how the increasing variety of identified ubiquitin signals are transduced will allow us to develop a deeper mechanistic perception of how the multiple factors come together to faithfully propagate genomic information. Here, we discuss these and additional conceptual challenges that are currently under study toward understanding how ubiquitin governs cell cycle regulation.
© 2017 Gilberto and Peter.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

