Characterization of transcriptional modules related to fibrosing-NAFLD progression
- PMID: 28684781
- PMCID: PMC5500537
- DOI: 10.1038/s41598-017-05044-2
Characterization of transcriptional modules related to fibrosing-NAFLD progression
Abstract
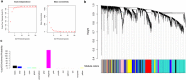
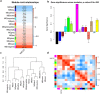
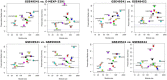
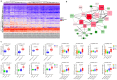
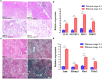
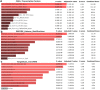
Based on the severity of liver fibrosis, low or high-risk profile of developing end-stage liver disease was present in nonalcoholic fatty liver disease (NAFLD). However, the mechanisms inducing transition from mild to advanced NAFLD are still elusive. We performed a system-level study on fibrosing-NAFLD by weighted gene co-expression network analysis (WGCNA) to identify significant modules in the network, and followed by functional and pathway enrichment analyses. Moreover, hub genes in the module were analyzed by network feature selection. As a result, fourteen distinct gene modules were identified, and seven modules showed significant associations with the status of NAFLD. Module preservation analysis confirmed that these modules can also be found in diverse independent datasets. After network feature analysis, the magenta module demonstrated a remarkably correlation with NAFLD fibrosis. The top hub genes with high connectivity or gene significance in the module were ultimately determined, including LUM, THBS2, FBN1 and EFEMP1. These genes were further verified in clinical samples. Finally, the potential regulators of magenta module were characterized. These findings highlighted a module and affiliated genes as playing important roles in the regulation of fibrosis in NAFLD, which may point to potential targets for therapeutic interventions.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Hashimoto E, Tokushige K, Ludwig J. Diagnosis and classification of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis: Current concepts and remaining challenges. Hepatology research: the official journal of the Japan Society of Hepatology. 2015;45:20–28. doi: 10.1111/hepr.12333. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

