Genomic and phenotypic analysis of Vavilov's historic landraces reveals the impact of environment and genomic islands of agronomic traits
- PMID: 28684880
- PMCID: PMC5500531
- DOI: 10.1038/s41598-017-05087-5
Genomic and phenotypic analysis of Vavilov's historic landraces reveals the impact of environment and genomic islands of agronomic traits
Abstract
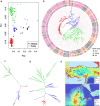
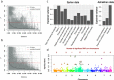
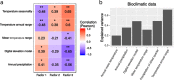
The Vavilov Institute of Plant Genetic Resources (VIR), in St. Petersburg, Russia, houses a unique genebank, with historical collections of landraces. When they were collected, the geographical distribution and genetic diversity of most crops closely reflected their historical patterns of cultivation established over the preceding millennia. We employed a combination of genomics, computational biology and phenotyping to characterize VIR's 147 chickpea accessions from Turkey and Ethiopia, representing chickpea's center of origin and a major location of secondary diversity. Genotyping by sequencing identified 14,059 segregating polymorphisms and genome-wide association studies revealed 28 GWAS hits in potential candidate genes likely to affect traits of agricultural importance. The proportion of polymorphisms shared among accessions is a strong predictor of phenotypic resemblance, and of environmental similarity between historical sampling sites. We found that 20 out of 28 polymorphisms, associated with multiple traits, including days to maturity, plant phenology, and yield-related traits such as pod number, localized to chromosome 4. We hypothesize that selection and introgression via inadvertent hybridization between more and less advanced morphotypes might have resulted in agricultural improvement genes being aggregated to genomic 'agro islands', and in genotype-to-phenotype relationships resembling widespread pleiotropy.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Genomic Analysis of Vavilov's Historic Chickpea Landraces Reveals Footprints of Environmental and Human Selection.Int J Mol Sci. 2020 May 31;21(11):3952. doi: 10.3390/ijms21113952. Int J Mol Sci. 2020. PMID: 32486400 Free PMC article.
-
Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits.Nat Genet. 2019 May;51(5):857-864. doi: 10.1038/s41588-019-0401-3. Epub 2019 Apr 29. Nat Genet. 2019. PMID: 31036963
-
Genetic dissection of plant growth habit in chickpea.Funct Integr Genomics. 2017 Nov;17(6):711-723. doi: 10.1007/s10142-017-0566-8. Epub 2017 Jun 9. Funct Integr Genomics. 2017. PMID: 28600722
-
Current advances in chickpea genomics: applications and future perspectives.Plant Cell Rep. 2018 Jul;37(7):947-965. doi: 10.1007/s00299-018-2305-6. Epub 2018 Jun 2. Plant Cell Rep. 2018. PMID: 29860584 Review.
-
Crop genome-wide association study: a harvest of biological relevance.Plant J. 2019 Jan;97(1):8-18. doi: 10.1111/tpj.14139. Epub 2018 Dec 17. Plant J. 2019. PMID: 30368955 Review.
Cited by
-
A Pipeline for Classifying Deleterious Coding Mutations in Agricultural Plants.Front Plant Sci. 2018 Nov 28;9:1734. doi: 10.3389/fpls.2018.01734. eCollection 2018. Front Plant Sci. 2018. PMID: 30546376 Free PMC article.
-
Toward the sequence-based breeding in legumes in the post-genome sequencing era.Theor Appl Genet. 2019 Mar;132(3):797-816. doi: 10.1007/s00122-018-3252-x. Epub 2018 Dec 17. Theor Appl Genet. 2019. PMID: 30560464 Free PMC article. Review.
-
Global-level population genomics reveals differential effects of geography and phylogeny on horizontal gene transfer in soil bacteria.Proc Natl Acad Sci U S A. 2019 Jul 23;116(30):15200-15209. doi: 10.1073/pnas.1900056116. Epub 2019 Jul 8. Proc Natl Acad Sci U S A. 2019. PMID: 31285337 Free PMC article.
-
Exploring the legacy of Central European historical winter wheat landraces.Sci Rep. 2021 Dec 13;11(1):23915. doi: 10.1038/s41598-021-03261-4. Sci Rep. 2021. PMID: 34903761 Free PMC article.
-
High-quality genome assembly and pan-genome studies facilitate genetic discovery in mung bean and its improvement.Plant Commun. 2022 Nov 14;3(6):100352. doi: 10.1016/j.xplc.2022.100352. Epub 2022 Jun 26. Plant Commun. 2022. PMID: 35752938 Free PMC article.
References
-
- Urruty N, Tailliez-Lefebvre D, Huyghe C. Stability, robustness, vulnerability and resilience of agricultural systems. A review. Agron. Sustain. Dev. 2016;36:1–15. doi: 10.1007/s13593-015-0347-5. - DOI
-
- Redden, R. J. & Berger, J. D. In Chickpea breeding and management (ed. Yadav, S. S., Redden, R. J., Chen, W. & Sharma, B.) 1–13 (CABI, 2007).
-
- Janick, J. N I Vavilov: Plant Geographer, Geneticist, Martyr of Science. HortScience a Publ. Am. Soc. Hortic. Sci. 50 (2014).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

