Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy
- PMID: 28685160
- PMCID: PMC5495776
- DOI: 10.1038/s41523-017-0028-4
Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy
Abstract
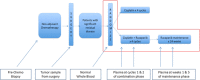
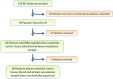
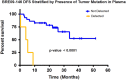
Next-generation sequencing to detect circulating tumor DNA is a minimally invasive method for tumor genotyping and monitoring therapeutic response. The majority of studies have focused on detecting circulating tumor DNA from patients with metastatic disease. Herein, we tested whether circulating tumor DNA could be used as a biomarker to predict relapse in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. In this study, we analyzed samples from 38 early-stage triple-negative breast cancer patients with matched tumor, blood, and plasma. Extracted DNA underwent library preparation and amplification using the Oncomine Research Panel consisting of 134 cancer genes, followed by high-coverage sequencing and bioinformatics. We detected high-quality somatic mutations from primary tumors in 33 of 38 patients. TP53 mutations were the most prevalent (82%) followed by PIK3CA (16%). Of the 33 patients who had a mutation identified in their primary tumor, we were able to detect circulating tumor DNA mutations in the plasma of four patients (three TP53 mutations, one AKT1 mutation, one CDKN2A mutation). All four patients had recurrence of their disease (100% specificity), but sensitivity was limited to detecting only 4 of 13 patients who clinically relapsed (31% sensitivity). Notably, all four patients had a rapid recurrence (0.3, 4.0, 5.3, and 8.9 months). Patients with detectable circulating tumor DNA had an inferior disease free survival (p < 0.0001; median disease-free survival: 4.6 mos. vs. not reached; hazard ratio = 12.6, 95% confidence interval: 3.06-52.2). Our study shows that next-generation circulating tumor DNA sequencing of triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy can predict recurrence with high specificity, but moderate sensitivity. For those patients where circulating tumor DNA is detected, recurrence is rapid.
Conflict of interest statement
D.B. and C.S. are employees at Thermo Fisher Scientific. The remaining authors declare no competing financial interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

