Reorganization of gene network for degradation of polycyclic aromatic hydrocarbons (PAHs) in Pseudomonas aeruginosa PAO1 under several conditions
- PMID: 28685384
- PMCID: PMC5655620
- DOI: 10.1007/s13353-017-0402-9
Reorganization of gene network for degradation of polycyclic aromatic hydrocarbons (PAHs) in Pseudomonas aeruginosa PAO1 under several conditions
Abstract
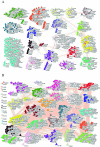
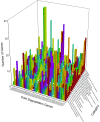
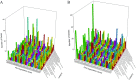
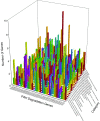
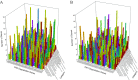
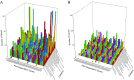
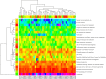
Although polycyclic aromatic hydrocarbons (PAHs) are harmful to human health, their elimination from the environment is not easy. Biodegradation of PAHs is promising since many bacteria have the ability to use hydrocarbons as their sole carbon and energy sources for growth. Of various microorganisms that can degrade PAHs, Pseudomonas aeruginosa is particularly important, not only because it causes a series of diseases including infection in cystic fibrosis patients, but also because it is a model bacterium in various studies. The genes that are responsible for degrading PAHs have been identified in P. aeruginosa, however, no gene acts alone as various stresses often initiate different metabolic pathways, quorum sensing, biofilm formation, antibiotic tolerance, etc. Therefore, it is important to study how PAH degradation genes behave under different conditions. In this study, we apply network analysis to investigating how 46 PAH degradation genes reorganized among 5549 genes in P. aeruginosa PAO1 under nine different conditions using publicly available gene coexpression data from GEO. The results provide six aspects of novelties: (i) comparing the number of gene clusters before and after stresses, (ii) comparing the membership in each gene cluster before and after stresses, (iii) defining which gene changed its membership together with PAH degradation genes before and after stresses, (iv) classifying membership-changed-genes in terms of category in Pseudomonas Genome Database, (v) postulating unknown gene's function, and (vi) proposing new mechanisms for genes of interests. This study can shed light on understanding of cooperative mechanisms of PAH degradation from the level of entire genes in an organism, and paves the way to conduct the similar studies on other genes.
Keywords: Bioinformatics; Network; Polycyclic aromatic hydrocarbon degradation gene; Pseudomonas Aeruginosa.
Conflict of interest statement
Funding
This study was partly supported by research grants from National Natural Science Foundation of China (31460296 and 31560315), and Special Funds for Building of Guangxi Talent Highland.
Conflict of interest
Authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures





Similar articles
-
Involvement of quorum sensing genes in biofilm development and degradation of polycyclic aromatic hydrocarbons by a marine bacterium Pseudomonas aeruginosa N6P6.Appl Microbiol Biotechnol. 2015 Dec;99(23):10283-97. doi: 10.1007/s00253-015-6868-7. Epub 2015 Aug 7. Appl Microbiol Biotechnol. 2015. PMID: 26245683
-
Whole genome characterization and phenanthrene catabolic pathway of a biofilm forming marine bacterium Pseudomonas aeruginosa PFL-P1.Ecotoxicol Environ Saf. 2020 Dec 15;206:111087. doi: 10.1016/j.ecoenv.2020.111087. Epub 2020 Aug 29. Ecotoxicol Environ Saf. 2020. PMID: 32871516
-
Multiple Roles for Two Efflux Pumps in the Polycyclic Aromatic Hydrocarbon-Degrading Pseudomonas putida Strain B6-2 (DSM 28064).Appl Environ Microbiol. 2017 Dec 1;83(24):e01882-17. doi: 10.1128/AEM.01882-17. Print 2017 Dec 15. Appl Environ Microbiol. 2017. PMID: 29030440 Free PMC article.
-
Biocatalytic Potential of Pseudomonas Species in the Degradation of Polycyclic Aromatic Hydrocarbons.J Basic Microbiol. 2025 Feb;65(2):e2400448. doi: 10.1002/jobm.202400448. Epub 2024 Oct 29. J Basic Microbiol. 2025. PMID: 39468883 Review.
-
A comprehensive review of metabolic and genomic aspects of PAH-degradation.Arch Microbiol. 2020 Oct;202(8):2033-2058. doi: 10.1007/s00203-020-01929-5. Epub 2020 Jun 6. Arch Microbiol. 2020. PMID: 32506150 Review.
Cited by
-
Microbial Consortia and Mixed Plastic Waste: Pangenomic Analysis Reveals Potential for Degradation of Multiple Plastic Types via Previously Identified PET Degrading Bacteria.Int J Mol Sci. 2022 May 17;23(10):5612. doi: 10.3390/ijms23105612. Int J Mol Sci. 2022. PMID: 35628419 Free PMC article.
-
Whole genome sequence analysis of Cupriavidus campinensis S14E4C, a heavy metal resistant bacterium.Mol Biol Rep. 2020 May;47(5):3973-3985. doi: 10.1007/s11033-020-05490-8. Epub 2020 May 13. Mol Biol Rep. 2020. PMID: 32406019 Free PMC article.
-
Bacteria, Fungi and Microalgae for the Bioremediation of Marine Sediments Contaminated by Petroleum Hydrocarbons in the Omics Era.Microorganisms. 2021 Aug 10;9(8):1695. doi: 10.3390/microorganisms9081695. Microorganisms. 2021. PMID: 34442774 Free PMC article. Review.
-
Current research on simultaneous oxidation of aliphatic and aromatic hydrocarbons by bacteria of genus Pseudomonas.Folia Microbiol (Praha). 2022 Aug;67(4):591-604. doi: 10.1007/s12223-022-00966-5. Epub 2022 Mar 22. Folia Microbiol (Praha). 2022. PMID: 35318574 Review.
-
Differential Expression and PAH Degradation: What Burkholderia vietnamiensis G4 Can Tell Us?Int J Microbiol. 2020 Aug 27;2020:8831331. doi: 10.1155/2020/8831331. eCollection 2020. Int J Microbiol. 2020. PMID: 32908529 Free PMC article.
References
-
- Balashova NV, Stolz A, Knackmuss HJ, Kosheleva IA, Naumov AV, Boronin AM. Purification and characterization of a salicylate hydroxylase involved in 1-hydroxy-2-naphthoic acid hydroxylation from the naphthalene and phenanthrene-degrading bacterial strain Pseudomonas putida BS202-P1. Biodegradation. 2001;12:179–188. doi: 10.1023/A:1013126723719. - DOI - PubMed
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

