Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction
- PMID: 28685770
- PMCID: PMC5539352
- DOI: 10.1038/cr.2017.88
Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction
Abstract
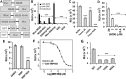
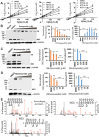
Recent outbreaks of Zika virus (ZIKV) highlight an urgent need for therapeutics. The protease complex NS2B-NS3 plays essential roles during flaviviral polyprotein processing, and thus represents an attractive drug target. Here, we developed a split luciferase complementation-based high-throughput screening assay to identify orthosteric inhibitors that directly target flavivirus NS2B-NS3 interactions. By screening a total of 2 816 approved and investigational drugs, we identified three potent candidates, temoporfin, niclosamide, and nitazoxanide, as flavivirus NS2B-NS3 interaction inhibitors with nanomolar potencies. Significantly, the most potent compound, temoporfin, not only inhibited ZIKV replication in human placental and neural progenitor cells, but also prevented ZIKV-induced viremia and mortality in mouse models. Structural docking suggests that temoporfin potentially binds NS3 pockets that hold critical NS2B residues, thus inhibiting flaviviral polyprotein processing in a non-competitive manner. As these drugs have already been approved for clinical use in other indications either in the USA or other countries, they represent promising and easily developed therapies for the management of infections by ZIKV and other flaviviruses.
Figures






References
-
- Calvet G, Aguiar RS, Melo AS, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis 2016; 16:653–660. - PubMed
-
- Chen LH, Hamer DH. Zika Virus: rapid spread in the western hemisphere. Ann Intern Med 2016; 164:613–615. - PubMed
-
- Ma W, Li S, Ma S, et al. Zika virus causes testis damage and leads to male infertility in mice. Cell 2016; 167:1511–1524. e15. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

