KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans
- PMID: 28685972
- PMCID: PMC5691316
- DOI: 10.1002/iid3.178
KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans
Abstract
Introduction: KIR2DS5 is an activating human NK cell receptor of lineage III KIR. These include both inhibitory KIR2DL1, 2 and 3 and activating KIR2DS1 that recognize either the C1 or C2 epitope of HLA-C. In Europeans KIR2DS5 is essentially monomorphic, with KIR2DS5*002 being predominant. Pioneering investigations showed that KIR2DS5*002 has activating potential, but cannot recognize HLA-A, -B, or -C. Subsequent studies have shown that KIR2DS5 is highly polymorphic in Africans, and that KIR2DS5*006 protects pregnant Ugandan women from preeclampsia. Because inhibitory C2-specific KIR2DL1 correlates with preeclampsia, whereas activating C2-specific KIR2DS1 protects, this association pointed to KIR2DS5*006 being an activating C2-specific receptor. To test this hypothesis we made KIR-Fc fusion proteins from all ten KIR2DS5 allotypes and tested their binding to a representative set of HLA-A, -B and -C allotypes.
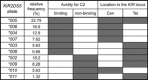
Results: Six African-specific KIR2DS5 bound to C2+ HLA-C but not to other HLA class I. Their avidity for C2 is ∼20% that of C2-specific KIR2DL1 and ∼40% that of C2-specific KIR2DS1. Among the African C2 receptors is KIR2DS5*006, which protected a cohort of pregnant Ugandans from pre-eclampsia. Three African KIR2DS5 allotypes and KIR2DS5*002, bound no HLA-A, -B or -C. As a group the C2-binding KIR2DS5 allotypes protect against pre-eclampsia compared to the non-binding KIR2DS5 allotypes. Natural substitutions that contribute to loss or reduction of C2 receptor function are at positions 127, 158, and 176 in the D2 domain.
Conclusions: KIR2DS5*005 has the KIR2DS5 consensus sequence, is the only allele found at both centromeric and telomeric locations of KIR2DS5, and is likely the common ancestor of all KIR2DS5 alleles. That KIR2DS5*005 has C2 receptor activity, points to KIR2DS5*002, and other allotypes lacking C2 receptor function, being products of attenuation, a characteristic feature of most KIR B haplotype genes. Alleles encoding attenuated and active KIR2DS5 are present in both centromeric and telomeric locations.
Keywords: Africa; HLA-C; KIR; NK cells; pregnancy.
© 2017 The Authors. Immunity, Inflammation and Disease Published by John Wiley & Sons Ltd.
Figures


Similar articles
-
Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I.J Immunol. 2010 Oct 1;185(7):4233-7. doi: 10.4049/jimmunol.1001951. Epub 2010 Aug 27. J Immunol. 2010. PMID: 20802150 Free PMC article.
-
Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes.J Immunol. 2015 Oct 1;195(7):3160-70. doi: 10.4049/jimmunol.1501358. Epub 2015 Aug 26. J Immunol. 2015. PMID: 26311903 Free PMC article.
-
Novel Approach to Cell Surface Discrimination Between KIR2DL1 Subtypes and KIR2DS1 Identifies Hierarchies in NK Repertoire, Education, and Tolerance.Front Immunol. 2019 Apr 5;10:734. doi: 10.3389/fimmu.2019.00734. eCollection 2019. Front Immunol. 2019. PMID: 31024561 Free PMC article.
-
KIR specificity and avidity of standard and unusual C1, C2, Bw4, Bw6 and A3/11 amino acid motifs at entire HLA:KIR interface between NK and target cells, the functional and evolutionary classification of HLA class I molecules.Int J Immunogenet. 2019 Aug;46(4):217-231. doi: 10.1111/iji.12433. Epub 2019 Jun 18. Int J Immunogenet. 2019. PMID: 31210416 Review.
-
Two to Tango: Co-evolution of Hominid Natural Killer Cell Receptors and MHC.Front Immunol. 2019 Feb 19;10:177. doi: 10.3389/fimmu.2019.00177. eCollection 2019. Front Immunol. 2019. PMID: 30837985 Free PMC article. Review.
Cited by
-
Correlation between human leukocyte antigen ligands and killer cell immunoglobulin-like receptors in aplastic anemia patients from Shaanxi Han.Immunogenetics. 2023 Oct;75(5):445-454. doi: 10.1007/s00251-023-01316-6. Epub 2023 Aug 17. Immunogenetics. 2023. PMID: 37592108
-
qKAT: Quantitative Semi-automated Typing of Killer-cell Immunoglobulin-like Receptor Genes.J Vis Exp. 2019 Mar 6;(145):10.3791/58646. doi: 10.3791/58646. J Vis Exp. 2019. PMID: 30907867 Free PMC article.
-
Influence of Fetomaternal Microchimerism on Maternal NK Cell Reactivity against the Child's Leukemic Blasts.Biomedicines. 2022 Mar 4;10(3):603. doi: 10.3390/biomedicines10030603. Biomedicines. 2022. PMID: 35327405 Free PMC article.
-
Uterine Natural Killer Cells: A Rising Star in Human Pregnancy Regulation.Front Immunol. 2022 Jun 1;13:918550. doi: 10.3389/fimmu.2022.918550. eCollection 2022. Front Immunol. 2022. PMID: 35720413 Free PMC article. Review.
-
Coexistence of inhibitory and activating killer-cell immunoglobulin-like receptors to the same cognate HLA-C2 and Bw4 ligands confer breast cancer risk.Sci Rep. 2021 Apr 12;11(1):7932. doi: 10.1038/s41598-021-86964-y. Sci Rep. 2021. PMID: 33846431 Free PMC article.
References
-
- Winter, C. C. , and Long E. O.. 1997. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA‐C allotypes. J. Immunol. 158:4026–4028. - PubMed
-
- Moesta, A. K. , Norman P. J., Yawata M., Yawata N., Gleimer M., and Parham P.. 2008. Synergistic polymorphism at two positions distal to the ligand‐binding site makes KIR2DL2 a stronger receptor for HLA‐C than KIR2 DL3. J. Immunol. 180:3969–3979. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

