Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity
- PMID: 28686870
- PMCID: PMC5642297
- DOI: 10.1016/j.stem.2017.06.014
Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity
Abstract
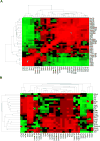
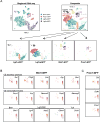
Several cell populations have been reported to possess intestinal stem cell (ISC) activity during homeostasis and injury-induced regeneration. Here, we explored inter-relationships between putative mouse ISC populations by comparative RNA-sequencing (RNA-seq). The transcriptomes of multiple cycling ISC populations closely resembled Lgr5+ ISCs, the most well-defined ISC pool, but Bmi1-GFP+ cells were distinct and enriched for enteroendocrine (EE) markers, including Prox1. Prox1-GFP+ cells exhibited sustained clonogenic growth in vitro, and lineage-tracing of Prox1+ cells revealed long-lived clones during homeostasis and after radiation-induced injury in vivo. Single-cell mRNA-seq revealed two subsets of Prox1-GFP+ cells, one of which resembled mature EE cells while the other displayed low-level EE gene expression but co-expressed tuft cell markers, Lgr5 and Ascl2, reminiscent of label-retaining secretory progenitors. Our data suggest that the EE lineage, including mature EE cells, comprises a reservoir of homeostatic and injury-inducible ISCs, extending our understanding of cellular plasticity and stemness.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- Anchang B, Hart TD, Bendall SC, Qiu P, Bjornson Z, Linderman M, Nolan GP, Plevritis SK. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nature protocols. 2016;11:1264–1279. - PubMed
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;7165;449:1003–1007. - PubMed
-
- Bezencon C, Furholz A, Raymond F, Mansourian R, Metairon S, Le Coutre J, Damak S. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. The Journal of comparative neurology. 2008;509:514–525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 DK085535/DK/NIDDK NIH HHS/United States
- K08 DK100739/DK/NIDDK NIH HHS/United States
- R01 EB020527/EB/NIBIB NIH HHS/United States
- R01 DK084056/DK/NIDDK NIH HHS/United States
- U01 DK085507/DK/NIDDK NIH HHS/United States
- K08 DK096048/DK/NIDDK NIH HHS/United States
- U01 DK085527/DK/NIDDK NIH HHS/United States
- T32 DK007647/DK/NIDDK NIH HHS/United States
- U01 DK085551/DK/NIDDK NIH HHS/United States
- U01 DK085508/DK/NIDDK NIH HHS/United States
- U01 DK085525/DK/NIDDK NIH HHS/United States
- U24 DK085532/DK/NIDDK NIH HHS/United States
- R03 DK114656/DK/NIDDK NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- U01 DK085547/DK/NIDDK NIH HHS/United States
- K08 HL119553/HL/NHLBI NIH HHS/United States
- U01 DK085532/DK/NIDDK NIH HHS/United States
- R01 CA227517/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

