A global evolutionary and metabolic analysis of human obesity gene risk variants
- PMID: 28687331
- PMCID: PMC5558862
- DOI: 10.1016/j.gene.2017.07.002
A global evolutionary and metabolic analysis of human obesity gene risk variants
Abstract
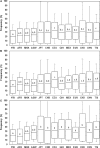
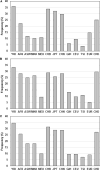
It is generally accepted that the selection of gene variants during human evolution optimized energy metabolism that now interacts with our obesogenic environment to increase the prevalence of obesity. The purpose of this study was to perform a global evolutionary and metabolic analysis of human obesity gene risk variants (110 human obesity genes with 127 nearest gene risk variants) identified using genome-wide association studies (GWAS) to enhance our knowledge of early and late genotypes. As a result of determining the mean frequency of these obesity gene risk variants in 13 available populations from around the world our results provide evidence for the early selection of ancestral risk variants (defined as selection before migration from Africa) and late selection of derived risk variants (defined as selection after migration from Africa). Our results also provide novel information for association of these obesity genes or encoded proteins with diverse metabolic pathways and other human diseases. The overall results indicate a significant differential evolutionary pattern for the selection of obesity gene ancestral and derived risk variants proposed to optimize energy metabolism in varying global environments and complex association with metabolic pathways and other human diseases. These results are consistent with obesity genes that encode proteins possessing a fundamental role in maintaining energy metabolism and survival during the course of human evolution.
Keywords: Environment; Evolution; Metabolism; Obesity; Variants.
Copyright © 2017. Published by Elsevier B.V.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

