Deficiency of RgpG Causes Major Defects in Cell Division and Biofilm Formation, and Deficiency of LytR-CpsA-Psr Family Proteins Leads to Accumulation of Cell Wall Antigens in Culture Medium by Streptococcus mutans
- PMID: 28687645
- PMCID: PMC5561293
- DOI: 10.1128/AEM.00928-17
Deficiency of RgpG Causes Major Defects in Cell Division and Biofilm Formation, and Deficiency of LytR-CpsA-Psr Family Proteins Leads to Accumulation of Cell Wall Antigens in Culture Medium by Streptococcus mutans
Abstract
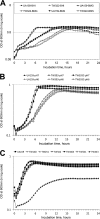
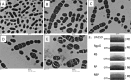
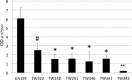
Streptococcus mutans is known to possess rhamnose-glucose polysaccharide (RGP), a major cell wall antigen. S. mutans strains deficient in rgpG, encoding the first enzyme of the RGP biosynthesis pathway, were constructed by allelic exchange. The rgpG deficiency had no effect on growth rate but caused major defects in cell division and altered cell morphology. Unlike the coccoid wild type, the rgpG mutant existed primarily in chains of swollen, "squarish" dividing cells. Deficiency of rgpG also causes significant reduction in biofilm formation (P < 0.01). Double and triple mutants with deficiency in brpA and/or psr, genes coding for the LytR-CpsA-Psr family proteins BrpA and Psr, which were previously shown to play important roles in cell envelope biogenesis, were constructed using the rgpG mutant. There were no major differences in growth rates between the wild-type strain and the rgpG brpA and rgpG psr double mutants, but the growth rate of the rgpG brpA psr triple mutant was reduced drastically (P < 0.001). Under transmission electron microscopy, both double mutants resembled the rgpG mutant, while the triple mutant existed as giant cells with multiple asymmetric septa. When analyzed by immunoblotting, the rgpG mutant displayed major reductions in cell wall antigens compared to the wild type, while little or no signal was detected with the double and triple mutants and the brpA and psr single mutants. These results suggest that RgpG in S. mutans plays a critical role in cell division and biofilm formation and that BrpA and Psr may be responsible for attachment of cell wall antigens to the cell envelope.IMPORTANCEStreptococcus mutans, a major etiological agent of human dental caries, produces rhamnose-glucose polysaccharide (RGP) as the major cell wall antigen. This study provides direct evidence that deficiency of RgpG, the first enzyme of the RGP biosynthesis pathway, caused major defects in cell division and morphology and reduced biofilm formation by S. mutans, indicative of a significant role of RGP in cell division and biofilm formation in S. mutans These results are novel not only in S. mutans, but also other streptococci that produce RGP. This study also shows that the LytR-CpsA-Psr family proteins BrpA and Psr in S. mutans are involved in attachment of RGP and probably other cell wall glycopolymers to the peptidoglycan. In addition, the results also suggest that BrpA and Psr may play a direct role in cell division and biofilm formation in S. mutans This study reveals new potential targets to develop anticaries therapeutics.
Keywords: BrpA; LCP proteins; Psr; Streptococcus mutans; TEM analysis; biofilm formation; cell division; cell wall antigens; dental caries; rhamnose-glucose polysaccharides.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
Psr is involved in regulation of glucan production, and double deficiency of BrpA and Psr is lethal in Streptococcus mutans.Microbiology (Reading). 2013 Mar;159(Pt 3):493-506. doi: 10.1099/mic.0.063032-0. Epub 2013 Jan 3. Microbiology (Reading). 2013. PMID: 23288544 Free PMC article.
-
Deficiency of BrpB causes major defects in cell division, stress responses and biofilm formation by Streptococcus mutans.Microbiology (Reading). 2014 Jan;160(Pt 1):67-78. doi: 10.1099/mic.0.072884-0. Epub 2013 Nov 4. Microbiology (Reading). 2014. PMID: 24190982 Free PMC article.
-
Disruption of l-Rhamnose Biosynthesis Results in Severe Growth Defects in Streptococcus mutans.J Bacteriol. 2020 Feb 25;202(6):e00728-19. doi: 10.1128/JB.00728-19. Print 2020 Feb 25. J Bacteriol. 2020. PMID: 31871035 Free PMC article.
-
LytR-CpsA-Psr Glycopolymer Transferases: Essential Bricks in Gram-Positive Bacterial Cell Wall Assembly.Int J Mol Sci. 2021 Jan 18;22(2):908. doi: 10.3390/ijms22020908. Int J Mol Sci. 2021. PMID: 33477538 Free PMC article. Review.
-
Polyketides/nonribosomal peptides from Streptococcus mutans and their ecological roles in dental biofilm.Mol Oral Microbiol. 2024 Oct;39(5):261-269. doi: 10.1111/omi.12451. Epub 2024 Jan 11. Mol Oral Microbiol. 2024. PMID: 38212261 Review.
Cited by
-
Multiple factors are involved in regulation of extracellular membrane vesicle biogenesis in Streptococcus mutans.Mol Oral Microbiol. 2021 Feb;36(1):12-24. doi: 10.1111/omi.12318. Epub 2020 Dec 3. Mol Oral Microbiol. 2021. PMID: 33040492 Free PMC article.
-
Structure and mechanism of biosynthesis of Streptococcus mutans cell wall polysaccharide.Nat Commun. 2025 Jan 22;16(1):954. doi: 10.1038/s41467-025-56205-1. Nat Commun. 2025. PMID: 39843487 Free PMC article.
-
Comparative Genomic Analysis of Streptococcus dysgalactiae subspecies dysgalactiae Isolated From Bovine Mastitis in China.Front Microbiol. 2021 Oct 22;12:751863. doi: 10.3389/fmicb.2021.751863. eCollection 2021. Front Microbiol. 2021. PMID: 34745056 Free PMC article.
-
Potential Risk of Spreading Resistance Genes within Extracellular-DNA-Dependent Biofilms of Streptococcus mutans in Response to Cell Envelope Stress Induced by Sub-MICs of Bacitracin.Appl Environ Microbiol. 2020 Aug 3;86(16):e00770-20. doi: 10.1128/AEM.00770-20. Print 2020 Aug 3. Appl Environ Microbiol. 2020. PMID: 32532873 Free PMC article.
-
Deficiency of BrpA in Streptococcus mutans reduces virulence in rat caries model.Mol Oral Microbiol. 2018 Oct;33(5):353-363. doi: 10.1111/omi.12230. Epub 2018 Jul 17. Mol Oral Microbiol. 2018. PMID: 29888871 Free PMC article.
References
-
- Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366. doi:10.1128/JB.01493-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

