Comparative Transcriptome Analysis Reveals Critical Function of Sucrose Metabolism Related-Enzymes in Starch Accumulation in the Storage Root of Sweet Potato
- PMID: 28690616
- PMCID: PMC5480015
- DOI: 10.3389/fpls.2017.00914
Comparative Transcriptome Analysis Reveals Critical Function of Sucrose Metabolism Related-Enzymes in Starch Accumulation in the Storage Root of Sweet Potato
Abstract
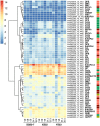
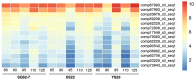
The starch properties of the storage root (SR) affect the quality of sweet potato (Ipomoea batatas (L.) Lam.). Although numerous studies have analyzed the accumulation and properties of starch in sweet potato SRs, the transcriptomic variation associated with starch properties in SR has not been quantified. In this study, we measured the starch and sugar contents and analyzed the transcriptome profiles of SRs harvested from sweet potatoes with high, medium, and extremely low starch contents, at five developmental stages [65, 80, 95, 110, and 125 days after transplanting (DAP)]. We found that differences in both water content and starch accumulation in the dry matter affect the starch content of SRs in different sweet potato genotypes. Based on transcriptome sequencing data, we assembled 112336 unigenes, and identified several differentially expressed genes (DEGs) involved in starch and sucrose metabolism, and revealed the transcriptional regulatory network controlling starch and sucrose metabolism in sweet potato SRs. Correlation analysis between expression patterns and starch and sugar contents suggested that the sugar-starch conversion steps catalyzed by sucrose synthase (SuSy) and UDP-glucose pyrophosphorylase (UGPase) may be essential for starch accumulation in the dry matter of SRs, and IbβFRUCT2, a vacuolar acid invertase, might also be a key regulator of starch content in the SRs. Our results provide valuable resources for future investigations aimed at deciphering the molecular mechanisms determining the starch properties of sweet potato SRs.
Keywords: RNA-seq; expression profile; starch; storage root; sucrose; sweet potato.
Figures






References
-
- Anwar N., Kikuchi A., Kumagai T., Watanabe K. N. (2009). Nucleotide sequence variation associated with β-amylase deficiency in the sweet potato Ipomoea batatas (L.) Lam. Breed. Sci. 59, 209–216. 10.1270/jsbbs.59.209 - DOI
-
- Cervantes-Flores J. C., Sosinski B., Pecota K. V., Mwanga R. O. M., Catignani G. L., Truong V. D., et al. (2011). Identification of quantitative trait loci for dry-matter, starch, and β-carotene content in sweetpotato. Mol. Breed. 28, 201–216. 10.1007/s11032-010-9474-5 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

