Drug Target Protein-Protein Interaction Networks: A Systematic Perspective
- PMID: 28691014
- PMCID: PMC5485489
- DOI: 10.1155/2017/1289259
Drug Target Protein-Protein Interaction Networks: A Systematic Perspective
Abstract
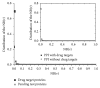
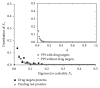
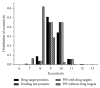
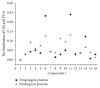
The identification and validation of drug targets are crucial in biomedical research and many studies have been conducted on analyzing drug target features for getting a better understanding on principles of their mechanisms. But most of them are based on either strong biological hypotheses or the chemical and physical properties of those targets separately. In this paper, we investigated three main ways to understand the functional biomolecules based on the topological features of drug targets. There are no significant differences between targets and common proteins in the protein-protein interactions network, indicating the drug targets are neither hub proteins which are dominant nor the bridge proteins. According to some special topological structures of the drug targets, there are significant differences between known targets and other proteins. Furthermore, the drug targets mainly belong to three typical communities based on their modularity. These topological features are helpful to understand how the drug targets work in the PPI network. Particularly, it is an alternative way to predict potential targets or extract nontargets to test a new drug target efficiently and economically. By this way, a drug target's homologue set containing 102 potential target proteins is predicted in the paper.
Figures





Similar articles
-
The analysis of the drug-targets based on the topological properties in the human protein-protein interaction network.J Drug Target. 2009 Aug;17(7):524-32. doi: 10.1080/10611860903046610. J Drug Target. 2009. PMID: 19530902
-
Are Topological Properties of Drug Targets Based on Protein-Protein Interaction Network Ready to Predict Potential Drug Targets?Comb Chem High Throughput Screen. 2016;19(2):109-20. doi: 10.2174/1386207319666151110122145. Comb Chem High Throughput Screen. 2016. PMID: 26552443
-
Analysis and identification of toxin targets by topological properties in protein-protein interaction network.J Theor Biol. 2014 May 21;349:82-91. doi: 10.1016/j.jtbi.2014.02.001. Epub 2014 Feb 7. J Theor Biol. 2014. PMID: 24512914
-
Some Remarks on Prediction of Drug-Target Interaction with Network Models.Curr Top Med Chem. 2017;17(21):2456-2468. doi: 10.2174/1568026617666170414145015. Curr Top Med Chem. 2017. PMID: 28413948 Review.
-
Network-based characterization of drug-regulated genes, drug targets, and toxicity.Methods. 2012 Aug;57(4):499-507. doi: 10.1016/j.ymeth.2012.06.003. Epub 2012 Jun 27. Methods. 2012. PMID: 22749929 Review.
Cited by
-
Identification of modules and functional analysis in CRC subtypes by integrated bioinformatics analysis.PLoS One. 2019 Aug 30;14(8):e0221772. doi: 10.1371/journal.pone.0221772. eCollection 2019. PLoS One. 2019. PMID: 31469863 Free PMC article.
-
Artificial intelligence in cancer target identification and drug discovery.Signal Transduct Target Ther. 2022 May 10;7(1):156. doi: 10.1038/s41392-022-00994-0. Signal Transduct Target Ther. 2022. PMID: 35538061 Free PMC article. Review.
-
Design and synthesis of a new orthogonally protected glutamic acid analog and its use in the preparation of high affinity polo-like kinase 1 polo-box domain - binding peptide macrocycles.Org Biomol Chem. 2021 Sep 22;19(36):7843-7854. doi: 10.1039/d1ob01120k. Org Biomol Chem. 2021. PMID: 34346472 Free PMC article.
-
TREAP: A New Topological Approach to Drug Target Inference.Biophys J. 2020 Dec 1;119(11):2290-2298. doi: 10.1016/j.bpj.2020.10.021. Epub 2020 Oct 29. Biophys J. 2020. PMID: 33129831 Free PMC article.
-
Patient-Specific Network for Personalized Breast Cancer Therapy with Multi-Omics Data.Entropy (Basel). 2021 Feb 11;23(2):225. doi: 10.3390/e23020225. Entropy (Basel). 2021. PMID: 33670375 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

