Generation and comparison of CRISPR-Cas9 and Cre-mediated genetically engineered mouse models of sarcoma
- PMID: 28691711
- PMCID: PMC5508130
- DOI: 10.1038/ncomms15999
Generation and comparison of CRISPR-Cas9 and Cre-mediated genetically engineered mouse models of sarcoma
Abstract
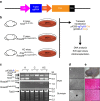
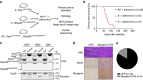
Genetically engineered mouse models that employ site-specific recombinase technology are important tools for cancer research but can be costly and time-consuming. The CRISPR-Cas9 system has been adapted to generate autochthonous tumours in mice, but how these tumours compare to tumours generated by conventional recombinase technology remains to be fully explored. Here we use CRISPR-Cas9 to generate multiple subtypes of primary sarcomas efficiently in wild type and genetically engineered mice. These data demonstrate that CRISPR-Cas9 can be used to generate multiple subtypes of soft tissue sarcomas in mice. Primary sarcomas generated with CRISPR-Cas9 and Cre recombinase technology had similar histology, growth kinetics, copy number variation and mutational load as assessed by whole exome sequencing. These results show that sarcomas generated with CRISPR-Cas9 technology are similar to sarcomas generated with conventional modelling techniques and suggest that CRISPR-Cas9 can be used to more rapidly generate genotypically and phenotypically similar cancers.
Conflict of interest statement
C.A.G. is a founder and scientific advisor to Element Genomics. The remaining authors declare no competing financial interests.
Figures




References
-
- Hanahan D., Wagner E. F. & Palmiter R. D. The origins of oncomice: a history of the first transgenic mice genetically engineered to develop cancer. Genes Dev. 21, 2258–2270 (2007). - PubMed
-
- Kirsch D. G. et al.. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat. Med. 13, 992–997 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

