Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia
- PMID: 28691929
- PMCID: PMC5531404
- DOI: 10.1172/JCI92069
Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia
Abstract
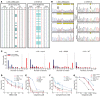
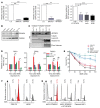
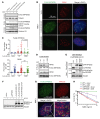
The WD40-containing E3 ubiquitin ligase RFWD3 has been recently linked to the repair of DNA damage by homologous recombination (HR). Here we have shown that an RFWD3 mutation within the WD40 domain is connected to the genetic disease Fanconi anemia (FA). An individual presented with congenital abnormalities characteristic of FA. Cells from the patient carrying the compound heterozygous mutations c.205_206dupCC and c.1916T>A in RFWD3 showed increased sensitivity to DNA interstrand cross-linking agents in terms of increased chromosomal breakage, reduced survival, and cell cycle arrest in G2 phase. The cellular phenotype was mirrored in genetically engineered human and avian cells by inactivation of RFWD3 or introduction of the patient-derived missense mutation, and the phenotype was rescued by expression of wild-type RFWD3 protein. HR was disrupted in RFWD3-mutant cells as a result of impaired relocation of mutant RFWD3 to chromatin and defective physical interaction with replication protein A. Rfwd3 knockout mice appear to have increased embryonic lethality, are subfertile, show ovarian and testicular atrophy, and have a reduced lifespan resembling that of other FA mouse models. Although RFWD3 mutations have thus far been detected in a single child with FA, we propose RFWD3 as an FA gene, FANCW, supported by cellular paradigm systems and an animal model.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

