Long non-coding RNA HOXA transcript at the distal tip as a biomarker for gastric cancer
- PMID: 28693275
- PMCID: PMC5494658
- DOI: 10.3892/ol.2017.6186
Long non-coding RNA HOXA transcript at the distal tip as a biomarker for gastric cancer
Abstract
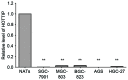
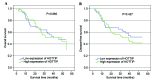
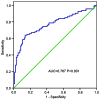
A long non-coding RNA named HOXA transcript at the distal tip (HOTTIP) has been reported to be significantly increased in several cancers, including hepatocellular cancer, pancreatic cancer and lung cancer. However, the clinical value of HOTTIP expression in gastric cancer remains unknown. The present study aimed to investigate HOTTIP expression levels in gastric cancer and to elucidate its clinical significance. Reverse transcription polymerase chain reaction was used to assess the expression level of HOTTIP in gastric cancer cell lines and tissues. In a cohort of 94 patients with gastric cancer, HOTTIP expression was significantly lower in cancer tissues compared with the normal adjacent tissues. In addition, receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of HOTTIP in gastric cancer, and the area under the ROC curve was 0.767. In conclusion, the results of the present study indicated that HOTTIP may be a predictive biomarker for gastric cancer.
Keywords: HOXA transcript at the distal tip; biomarker; gastric cancer; long non-coding RNA.
Figures

Similar articles
-
The HOTTIP (HOXA transcript at the distal tip) lncRNA: Review of oncogenic roles in human.Biomed Pharmacother. 2020 Jul;127:110158. doi: 10.1016/j.biopha.2020.110158. Epub 2020 Apr 23. Biomed Pharmacother. 2020. PMID: 32335298 Review.
-
HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression.Oncol Rep. 2016 Jun;35(6):3577-85. doi: 10.3892/or.2016.4743. Epub 2016 Apr 13. Oncol Rep. 2016. PMID: 27108607
-
Expression levels of long non-coding RNA HOXA distal transcript antisense RNA and metabotropic glutamate receptor 1 in pancreatic carcinoma, and their prognostic values.Oncol Lett. 2018 Jun;15(6):9464-9470. doi: 10.3892/ol.2018.8519. Epub 2018 Apr 17. Oncol Lett. 2018. PMID: 29844833 Free PMC article.
-
Investigation of the Use of Circulating Long Non-coding RNA HOXA Transcript at the Distal Tip (LncRNA HOTTIP) as a Biomarker in Breast Cancer.Cureus. 2023 Dec 6;15(12):e50019. doi: 10.7759/cureus.50019. eCollection 2023 Dec. Cureus. 2023. PMID: 38186456 Free PMC article.
-
Long Non-Coding RNA HOXA Transcript at the Distal Tip as a Putative Biomarker of Metastasis and Prognosis: a Meta-Analysis.Clin Lab. 2016 Nov 1;62(11):2091-2098. doi: 10.7754/Clin.Lab.2016.160224. Clin Lab. 2016. PMID: 28164662 Review.
Cited by
-
The long antisense non-coding RNA HOXA transcript at the distal tip (LncRNA HOTTIP) in health and disease: a comprehensive review and in silico analysis.Naunyn Schmiedebergs Arch Pharmacol. 2025 Jul 5. doi: 10.1007/s00210-025-04372-9. Online ahead of print. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 40616679 Review.
-
One stomach, two subtypes of carcinoma-the differences between distal and proximal gastric cancer.Gastroenterol Rep (Oxf). 2021 Nov 15;9(6):489-504. doi: 10.1093/gastro/goab050. eCollection 2021 Dec. Gastroenterol Rep (Oxf). 2021. PMID: 34925847 Free PMC article. Review.
-
Apoptotic Effects of Linum album Extracts on AGS Human Gastric Adenocarcinoma Cells and ZNF703 Oncogene Expression.Asian Pac J Cancer Prev. 2018 Oct 26;19(10):2911-2916. doi: 10.22034/APJCP.2018.19.10.2911. Asian Pac J Cancer Prev. 2018. PMID: 30362321 Free PMC article.
-
Prognostic value of long noncoding RNAs in gastric cancer: a meta-analysis.Onco Targets Ther. 2018 Aug 14;11:4877-4891. doi: 10.2147/OTT.S169823. eCollection 2018. Onco Targets Ther. 2018. PMID: 30147339 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
