Human presence impacts fungal diversity of inflated lunar/Mars analog habitat
- PMID: 28693587
- PMCID: PMC5504618
- DOI: 10.1186/s40168-017-0280-8
Human presence impacts fungal diversity of inflated lunar/Mars analog habitat
Abstract
Background: An inflatable lunar/Mars analog habitat (ILMAH), simulated closed system isolated by HEPA filtration, mimics International Space Station (ISS) conditions and future human habitation on other planets except for the exchange of air between outdoor and indoor environments. The ILMAH was primarily commissioned to measure physiological, psychological, and immunological characteristics of human inhabiting in isolation, but it was also available for other studies such as examining its microbiological aspects. Characterizing and understanding possible changes and succession of fungal species is of high importance since fungi are not only hazardous to inhabitants but also deteriorate the habitats. Observing the mycobiome changes in the presence of human will enable developing appropriate countermeasures with reference to crew health in a future closed habitat.
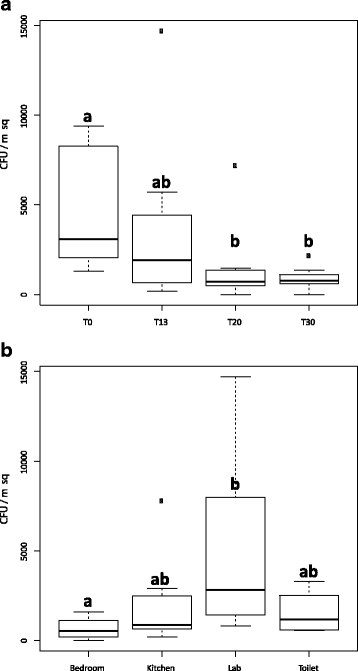
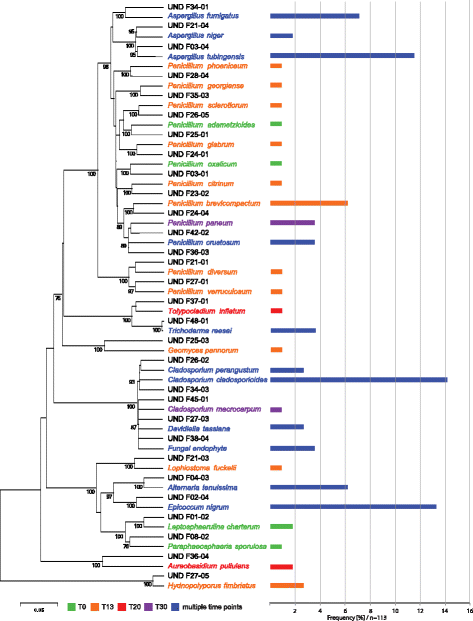
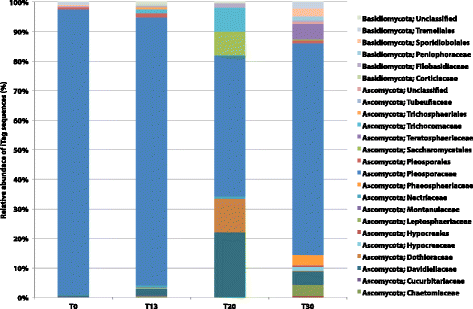
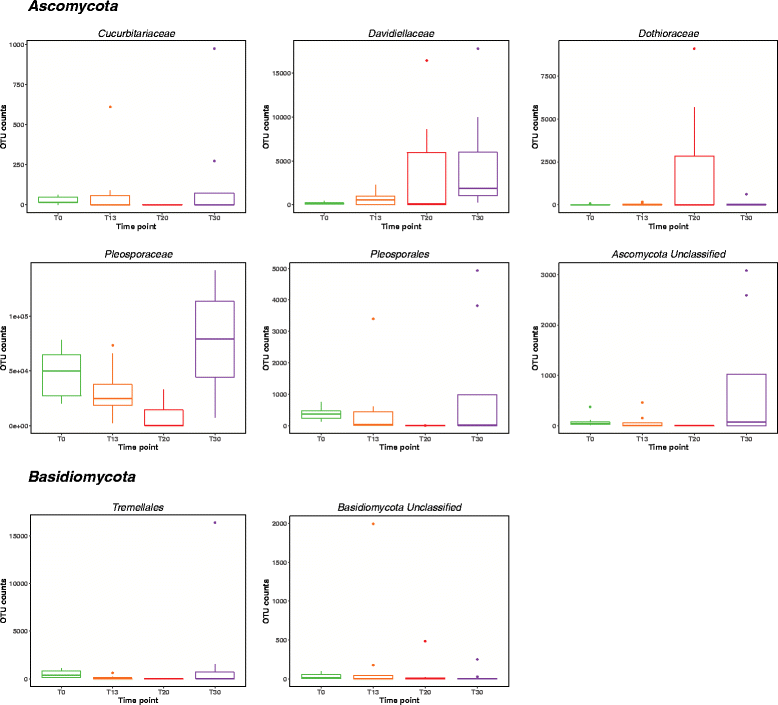
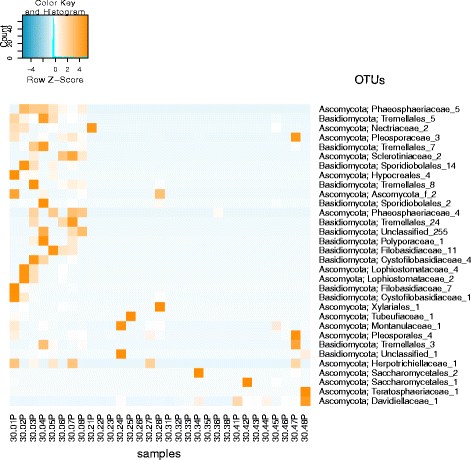
Results: Succession of fungi was characterized utilizing both traditional and state-of-the-art molecular techniques during the 30-day human occupation of the ILMAH. Surface samples were collected at various time points and locations to observe both the total and viable fungal populations of common environmental and opportunistic pathogenic species. To estimate the cultivable fungal population, potato dextrose agar plate counts method was utilized. The internal transcribed spacer region-based iTag Illumina sequencing was employed to measure the community structure and fluctuation of the mycobiome over time in various locations. Treatment of samples with propidium monoazide (PMA; a DNA intercalating dye for selective detection of viable microbial populations) had a significant effect on the microbial diversity compared to non-PMA-treated samples. Statistical analysis confirmed that viable fungal community structure changed (increase in diversity and decrease in fungal burden) over the occupation time. Samples collected at day 20 showed distinct fungal profiles from samples collected at any other time point (before or after). Viable fungal families like Davidiellaceae, Teratosphaeriaceae, Pleosporales, and Pleosporaceae were shown to increase during the occupation time.
Conclusions: The results of this study revealed that the overall fungal diversity in the closed habitat changed during human presence; therefore, it is crucial to properly maintain a closed habitat to preserve it from deteriorating and keep it safe for its inhabitants. Differences in community profiles were observed when statistically treated, especially of the mycobiome of samples collected at day 20. On a genus level Epiccocum, Alternaria, Pleosporales, Davidiella, and Cryptococcus showed increased abundance over the occupation time.
Keywords: Closed habitat; Mycobiome; Succession; Surface.
Figures



Similar articles
-
Microbial succession in an inflated lunar/Mars analog habitat during a 30-day human occupation.Microbiome. 2016 Jun 2;4(1):22. doi: 10.1186/s40168-016-0167-0. Microbiome. 2016. PMID: 27250991 Free PMC article.
-
Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces.Microbiome. 2019 Apr 8;7(1):50. doi: 10.1186/s40168-019-0666-x. Microbiome. 2019. PMID: 30955503 Free PMC article.
-
Surface fungal diversity and several mycotoxin-related genes' expression profiles during the Lunar Palace 365 experiment.Microbiome. 2022 Oct 12;10(1):169. doi: 10.1186/s40168-022-01350-8. Microbiome. 2022. PMID: 36224642 Free PMC article.
-
Dimensions of biodiversity in the Earth mycobiome.Nat Rev Microbiol. 2016 Jul;14(7):434-47. doi: 10.1038/nrmicro.2016.59. Nat Rev Microbiol. 2016. PMID: 27296482 Review.
-
Emerging Insights into the Occupational Mycobiome.Curr Allergy Asthma Rep. 2018 Sep 27;18(11):62. doi: 10.1007/s11882-018-0818-2. Curr Allergy Asthma Rep. 2018. PMID: 30259186 Free PMC article. Review.
Cited by
-
The International Space Station Environment Triggers Molecular Responses in Aspergillus niger.Front Microbiol. 2022 Jun 30;13:893071. doi: 10.3389/fmicb.2022.893071. eCollection 2022. Front Microbiol. 2022. PMID: 35847112 Free PMC article.
-
Microbiome in a ground-based analog cabin of China Space Station during a 50-day human occupation.ISME Commun. 2024 Jan 24;4(1):ycae013. doi: 10.1093/ismeco/ycae013. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38495633 Free PMC article.
-
Current Progression: Application of High-Throughput Sequencing Technique in Space Microbiology.Biomed Res Int. 2020 Jun 20;2020:4094191. doi: 10.1155/2020/4094191. eCollection 2020. Biomed Res Int. 2020. PMID: 32685480 Free PMC article. Review.
-
Genomic and morphological characterization of Knufia obscura isolated from the Mars 2020 spacecraft assembly facility.Sci Rep. 2024 May 28;14(1):12249. doi: 10.1038/s41598-024-61115-1. Sci Rep. 2024. PMID: 38806503 Free PMC article.
-
Genomic characterization and radiation tolerance of Naganishia kalamii sp. nov. and Cystobasidium onofrii sp. nov. from Mars 2020 mission assembly facilities.IMA Fungus. 2023 Aug 11;14(1):15. doi: 10.1186/s43008-023-00119-4. IMA Fungus. 2023. PMID: 37568226 Free PMC article.
References
-
- Price H, Baker J, Naderi F. A minimal architecture for human journeys to Mars. New Space. 2015;3(2):73–81. doi: 10.1089/space.2015.0018. - DOI
-
- Wilhite AW, Chai P. Plan B for U.S. Human Space Exploration Program. 2014.
-
- Swarmer TM, Anderson L, de León P: Performance review of a pressurized inflatable lunar habitat integrated with an electric rover and pressurized analog planetary suits during an initial ten day simulation. International Conference on Environmental Systems 2014.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

