Normal breast tissue DNA methylation differences at regulatory elements are associated with the cancer risk factor age
- PMID: 28693600
- PMCID: PMC5504720
- DOI: 10.1186/s13058-017-0873-y
Normal breast tissue DNA methylation differences at regulatory elements are associated with the cancer risk factor age
Abstract
Background: The underlying biological mechanisms through which epidemiologically defined breast cancer risk factors contribute to disease risk remain poorly understood. Identification of the molecular changes associated with cancer risk factors in normal tissues may aid in determining the earliest events of carcinogenesis and informing cancer prevention strategies.
Methods: Here we investigated the impact cancer risk factors have on the normal breast epigenome by analyzing DNA methylation genome-wide (Infinium 450 K array) in cancer-free women from the Susan G. Komen Tissue Bank (n = 100). We tested the relation of established breast cancer risk factors, age, body mass index, parity, and family history of disease, with DNA methylation adjusting for potential variation in cell-type proportions.
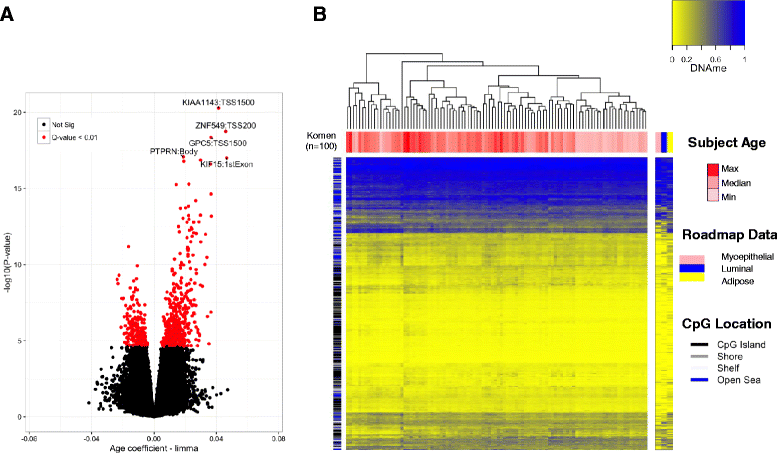
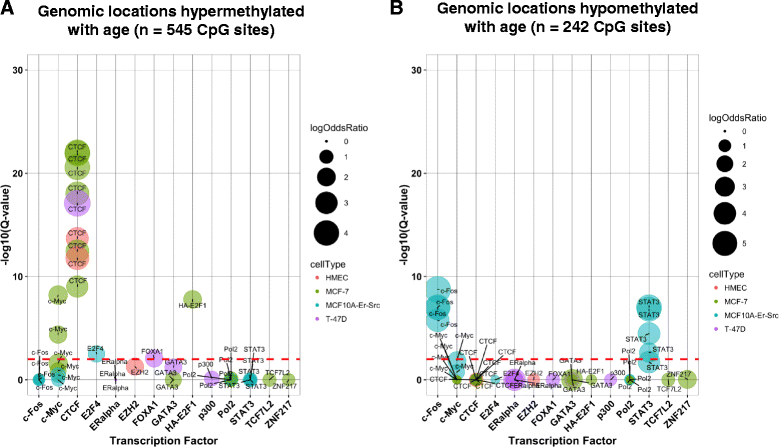
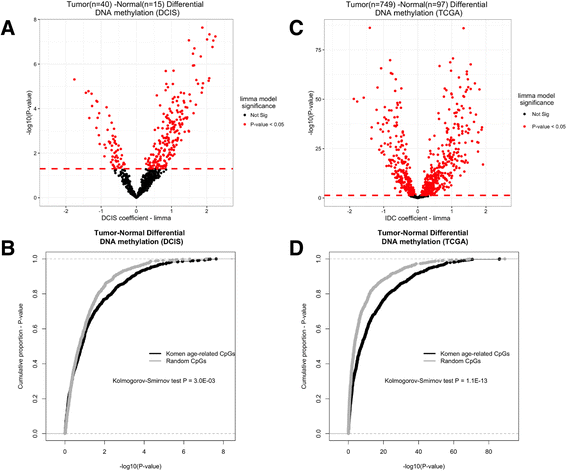
Results: We identified 787 cytosine-guanine dinucleotide (CpG) sites that demonstrated significant associations (Q value <0.01) with subject age. Notably, DNA methylation was not strongly associated with the other evaluated breast cancer risk factors. Age-related DNA methylation changes are primarily increases in methylation enriched at breast epithelial cell enhancer regions (P = 7.1E-20), and binding sites of chromatin remodelers (MYC and CTCF). We validated the age-related associations in two independent populations, using normal breast tissue samples (n = 18) and samples of normal tissue adjacent to tumor tissue (n = 97). The genomic regions classified as age-related were more likely to be regions altered in both pre-invasive (n = 40, P = 3.0E-03) and invasive breast tumors (n = 731, P = 1.1E-13).
Conclusions: DNA methylation changes with age occur at regulatory regions, and are further exacerbated in cancer, suggesting that age influences breast cancer risk in part through its contribution to epigenetic dysregulation in normal breast tissue.
Keywords: 5mC; Aging; Breast cancer; DNA methylation; Epigenetic drift; Epigenetics; Normal breast; Reference-free; Risk factors.
Conflict of interest statement
Ethics approval and consent to participate
Subject consent and approval from the Dartmouth College Institutional Review Board were obtained prior to the use of these tissues for research purposes (number 00023420). This work was performed in accordance with the ethical principles in the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Hamajima N, Hirose K, Tajima K, Rohan T, Calle EE, Heath CW, Jr, Coates RJ, Liff JM, Talamini R, Chantarakul N, et al. Alcohol, tobacco and breast cancer–collaborative reanalysis of individual data from 53 epidemiological studies, including 58,515 women with breast cancer and 95,067 women without the disease. Br J Cancer. 2002;87(11):1234–45. doi: 10.1038/sj.bjc.6600596. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

