High-efficiency non-mosaic CRISPR-mediated knock-in and indel mutation in F0 Xenopus
- PMID: 28694259
- PMCID: PMC5560047
- DOI: 10.1242/dev.152967
High-efficiency non-mosaic CRISPR-mediated knock-in and indel mutation in F0 Xenopus
Abstract
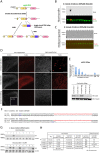
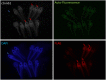
The revolution in CRISPR-mediated genome editing has enabled the mutation and insertion of virtually any DNA sequence, particularly in cell culture where selection can be used to recover relatively rare homologous recombination events. The efficient use of this technology in animal models still presents a number of challenges, including the time to establish mutant lines, mosaic gene editing in founder animals, and low homologous recombination rates. Here we report a method for CRISPR-mediated genome editing in Xenopus oocytes with homology-directed repair (HDR) that provides efficient non-mosaic targeted insertion of small DNA fragments (40-50 nucleotides) in 4.4-25.7% of F0 tadpoles, with germline transmission. For both CRISPR/Cas9-mediated HDR gene editing and indel mutation, the gene-edited F0 embryos are uniformly heterozygous, consistent with a mutation in only the maternal genome. In addition to efficient tagging of proteins in vivo, this HDR methodology will allow researchers to create patient-specific mutations for human disease modeling in Xenopus.
Keywords: CRISPR/Cas9; F0; Homology-directed repair (HDR); Non-mosaic; Xenopus laevis; Xenopus tropicalis.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

