Transcriptional Elongation Control of Hepatitis B Virus Covalently Closed Circular DNA Transcription by Super Elongation Complex and BRD4
- PMID: 28694331
- PMCID: PMC5599714
- DOI: 10.1128/MCB.00040-17
Transcriptional Elongation Control of Hepatitis B Virus Covalently Closed Circular DNA Transcription by Super Elongation Complex and BRD4
Abstract
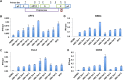
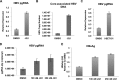
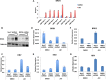
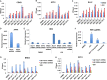
Chronic hepatitis B virus (HBV) infection can lead to liver cirrhosis and hepatocellular carcinoma. HBV reactivation during or after chemotherapy is a potentially fatal complication for cancer patients with chronic HBV infection. Transcription of HBV is a critical intermediate step of the HBV life cycle. However, factors controlling HBV transcription remain largely unknown. Here, we found that different P-TEFb complexes are involved in the transcription of the HBV viral genome. Both BRD4 and the super elongation complex (SEC) bind to the HBV genome. The treatment of bromodomain inhibitor JQ1 stimulates HBV transcription and increases the occupancy of BRD4 on the HBV genome, suggesting the bromodomain-independent recruitment of BRD4 to the HBV genome. JQ1 also leads to the increased binding of SEC to the HBV genome, and SEC is required for JQ1-induced HBV transcription. These findings reveal a novel mechanism by which the HBV genome hijacks the host P-TEFb-containing complexes to promote its own transcription. Our findings also point out an important clinical implication, that is, the potential risk of HBV reactivation during therapy with a BRD4 inhibitor, such as JQ1 or its analogues, which are a potential treatment for acute myeloid leukemia.
Keywords: BRD4; hepatitis B virus; super elongation complex; transcriptional elongation.
Copyright © 2017 American Society for Microbiology.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
