Analysis of positive and negative allosteric modulation in metabotropic glutamate receptors 4 and 5 with a dual ligand
- PMID: 28694498
- PMCID: PMC5504000
- DOI: 10.1038/s41598-017-05095-5
Analysis of positive and negative allosteric modulation in metabotropic glutamate receptors 4 and 5 with a dual ligand
Abstract
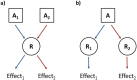
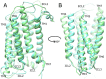
As class C GPCRs and regulators of synaptic activity, human metabotropic glutamate receptors (mGluRs) 4 and 5 are prime targets for allosteric modulation, with mGlu5 inhibition or mGlu4 stimulation potentially treating conditions like chronic pain and Parkinson's disease. As an allosteric modulator that can bind both receptors, 2-Methyl-6-(phenylethynyl)pyridine (MPEP) is able to negatively modulate mGlu5 or positively modulate mGlu4. At a structural level, how it elicits these responses and how mGluRs undergo activation is unclear. Here, we employ homology modelling and 30 µs of atomistic molecular dynamics (MD) simulations to probe allosteric conformational change in mGlu4 and mGlu5, with and without docked MPEP. Our results identify several structural differences between mGlu4 and mGlu5, as well as key differences responsible for MPEP-mediated positive and negative allosteric modulation, respectively. A novel mechanism of mGlu4 activation is revealed, which may apply to all mGluRs in general. This involves conformational changes in TM3, TM4 and TM5, separation of intracellular loop 2 (ICL2) from ICL1/ICL3, and destabilization of the ionic-lock. On the other hand, mGlu5 experiences little disturbance when MPEP binds, maintaining its inactive state with reduced conformational fluctuation. In addition, when MPEP is absent, a lipid molecule can enter the mGlu5 allosteric pocket.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
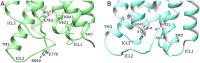


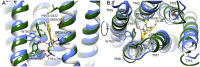



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

