Comparative Genomic Characterization of the Highly Persistent and Potentially Virulent Cronobacter sakazakii ST83, CC65 Strain H322 and Other ST83 Strains
- PMID: 28694793
- PMCID: PMC5483470
- DOI: 10.3389/fmicb.2017.01136
Comparative Genomic Characterization of the Highly Persistent and Potentially Virulent Cronobacter sakazakii ST83, CC65 Strain H322 and Other ST83 Strains
Abstract
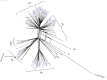
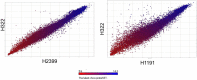
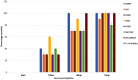
Cronobacter (C.) sakazakii is an opportunistic pathogen and has been associated with serious infections with high mortality rates predominantly in pre-term, low-birth weight and/or immune compromised neonates and infants. Infections have been epidemiologically linked to consumption of intrinsically and extrinsically contaminated lots of reconstituted powdered infant formula (PIF), thus contamination of such products is a challenging task for the PIF producing industry. We present the draft genome of C. sakazakii H322, a highly persistent sequence type (ST) 83, clonal complex (CC) 65, serotype O:7 strain obtained from a batch of non-released contaminated PIF product. The presence of this strain in the production environment was traced back more than 4 years. Whole genome sequencing (WGS) of this strain together with four more ST83 strains (PIF production environment-associated) confirmed a high degree of sequence homology among four of the five strains. Phylogenetic analysis using microarray (MA) and WGS data showed that the ST83 strains were highly phylogenetically related and MA showed that between 5 and 38 genes differed from one another in these strains. All strains possessed the pESA3-like virulence plasmid and one strain possessed a pESA2-like plasmid. In addition, a pCS1-like plasmid was also found. In order to assess the potential in vivo pathogenicity of the ST83 strains, each strain was subjected to infection studies using the recently developed zebrafish embryo model. Our results showed a high (90-100%) zebrafish mortality rate for all of these strains, suggesting a high risk for infections and illness in neonates potentially exposed to PIF contaminated with ST83 C. sakazakii strains. In summary, virulent ST83, CC65, serotype CsakO:7 strains, though rarely found intrinsically in PIF, can persist within a PIF manufacturing facility for years and potentially pose significant quality assurance challenges to the PIF manufacturing industry.
Keywords: Cronobacter; environmental microarray; persister; whole genome sequencing.
Figures


References
-
- Affymetrix (2014). Expression Analysis Technical Manual, with Specific Protocols for Use with the Hybridization, Wash, and Stain Kit. Available at: http://www.affymetrix.com/support/downloads/manuals/expression_analysis_...
-
- Chase H. R., Gopinath G. R., Gangiredla J., Patel I. R., Kothary M. H., Carter L., et al. (2016). Genome sequences of malonate-positive Cronobacter sakazakii serogroup O:2, sequence type 64 strains CDC 1121-73 and GK1025, isolated from human bronchial wash and a powdered infant formula manufacturing plant. Genome Announc. 4:e01072–16 10.1128/genomeA.01072-16 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

