Towards open-ended evolution in self-replicating molecular systems
- PMID: 28694865
- PMCID: PMC5496545
- DOI: 10.3762/bjoc.13.118
Towards open-ended evolution in self-replicating molecular systems
Abstract
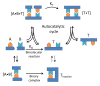
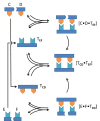
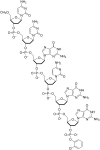
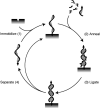
In this review we discuss systems of self-replicating molecules in the context of the origin of life and the synthesis of de novo life. One of the important aspects of life is the ability to reproduce and evolve continuously. In this review we consider some of the prerequisites for obtaining unbounded evolution of self-replicating molecules and describe some recent advances in this field. While evolution experiments involving self-replicating molecules have shown promising results, true open-ended evolution has not been realized so far. A full understanding of the requirements for open-ended evolution would provide a better understanding of how life could have emerged from molecular building blocks and what is needed to create a minimal form of life in the laboratory.
Keywords: autocatalysis; open-ended evolution; origin of life; self-replication; synthetic life.
Figures


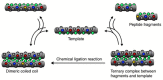
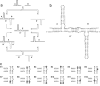

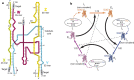
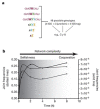
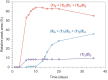
Similar articles
-
An Approach to the De Novo Synthesis of Life.Acc Chem Res. 2022 Jan 18;55(2):145-155. doi: 10.1021/acs.accounts.1c00534. Epub 2021 Dec 29. Acc Chem Res. 2022. PMID: 34964346 Free PMC article.
-
From self-replication to replicator systems en route to de novo life.Nat Rev Chem. 2020 Aug;4(8):386-403. doi: 10.1038/s41570-020-0196-x. Epub 2020 Jul 1. Nat Rev Chem. 2020. PMID: 37127968 Review.
-
Formal Definitions of Unbounded Evolution and Innovation Reveal Universal Mechanisms for Open-Ended Evolution in Dynamical Systems.Sci Rep. 2017 Apr 20;7(1):997. doi: 10.1038/s41598-017-00810-8. Sci Rep. 2017. PMID: 28428620 Free PMC article.
-
A theory for the origin of a self-replicating chemical system. I: Natural selection of the autogen from short, random oligomers.J Mol Evol. 1980 Dec;16(2):121-47. doi: 10.1007/BF01731582. J Mol Evol. 1980. PMID: 6162031
-
Self-replicating systems.Org Biomol Chem. 2016 May 4;14(18):4170-84. doi: 10.1039/c6ob00280c. Org Biomol Chem. 2016. PMID: 27086507 Review.
Cited by
-
An Optical Probe for Real-Time Monitoring of Self-Replicator Emergence and Distinguishing between Replicators.J Am Chem Soc. 2022 Feb 23;144(7):3074-3082. doi: 10.1021/jacs.1c11594. Epub 2022 Feb 9. J Am Chem Soc. 2022. PMID: 35139307 Free PMC article.
-
Molecular basis of Arginine and Lysine DNA sequence-dependent thermo-stability modulation.PLoS Comput Biol. 2022 Jan 10;18(1):e1009749. doi: 10.1371/journal.pcbi.1009749. eCollection 2022 Jan. PLoS Comput Biol. 2022. PMID: 35007284 Free PMC article.
-
The Future of Origin of Life Research: Bridging Decades-Old Divisions.Life (Basel). 2020 Feb 26;10(3):20. doi: 10.3390/life10030020. Life (Basel). 2020. PMID: 32110893 Free PMC article.
-
Evolution at the Origins of Life?Life (Basel). 2024 Jan 24;14(2):175. doi: 10.3390/life14020175. Life (Basel). 2024. PMID: 38398684 Free PMC article. Review.
-
Self-Reproduction and Darwinian Evolution in Autocatalytic Chemical Reaction Systems.Life (Basel). 2021 Apr 1;11(4):308. doi: 10.3390/life11040308. Life (Basel). 2021. PMID: 33916135 Free PMC article. Review.
References
-
- Kauffmann S A. At Home in the Universe. New York, USA: Oxford University Press; 1995.
-
- Darwin C. On the Origin of Species. New York, USA: D. Appleton and CO.; 1871.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
