Evaluation of a Probe-Based PCR-ELISA System for Simultaneous Semi Quantitative Detection and Genotyping of Human Cytomegalovirus (HCMV) Infection in Clinical Specimens
- PMID: 28694881
- PMCID: PMC5481617
- DOI: 10.2174/1874285801711010083
Evaluation of a Probe-Based PCR-ELISA System for Simultaneous Semi Quantitative Detection and Genotyping of Human Cytomegalovirus (HCMV) Infection in Clinical Specimens
Abstract
Background: Human cytomegalovirus (HCMV) is a common opportunistic pathogen that causes serious complications in immunosuppressed patients and infected newborns. In this study, PCR-ELISA was optimized for semi-quantitative detection of infection in clinical specimens and simultaneous genotyping of glycoprotein B for 4 major genotypes, due to its significance.
Method: During DIG-labeling PCR, a pair of primers amplifies a fragment of variable region of the glycoprotein B encoding sequence. Under optimized conditions, labeled Target amplicons hybridize to biotinated specific probes and are detected in an ELISA system.
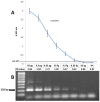
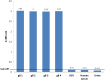
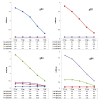
Results: PCR-ELISA system showed specific performance with detection limit of approximately 100 copies of CMV DNA. The linear correlation was observed between the PCR-ELISA results (OD) and logarithmic scale of CMV (r=0.979). Repeatability of PCR-ELISA detection system for intra-assay and inter-assay was evaluated for negative and positive samples. In optimized conditions of hybridization, differentiation between genotypes of glycoprotein B was feasible using genotype-specific probes in PCR-ELISA genotyping system. In comparison with sequencing method, genotyping system was confirmed with kappa index of 1.
Conclusion: PCR-ELISA is proposed as an applicable and reliable technique for semi-quantitative diagnosis and typing of the infection. This technique is flexible to apply in a variety of molecular fields.
Keywords: Genotyping; Glycoprotein B; Human cytomegalovirus; PCR-ELISA; Semi-quantitative detection.
Figures
References
-
- Knipe D., Howley P. Fields virology. 5th ed. USA: Lippincott Williams and Wilkins; 2007. pp. 2703–2756. Cytomegaloviruses.
LinkOut - more resources
Full Text Sources
Other Literature Sources
