Population genomics of picophytoplankton unveils novel chromosome hypervariability
- PMID: 28695208
- PMCID: PMC5498103
- DOI: 10.1126/sciadv.1700239
Population genomics of picophytoplankton unveils novel chromosome hypervariability
Abstract
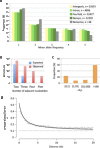
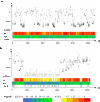
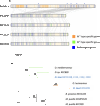
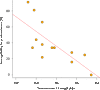
Tiny photosynthetic microorganisms that form the picoplankton (between 0.3 and 3 μm in diameter) are at the base of the food web in many marine ecosystems, and their adaptability to environmental change hinges on standing genetic variation. Although the genomic and phenotypic diversity of the bacterial component of the oceans has been intensively studied, little is known about the genomic and phenotypic diversity within each of the diverse eukaryotic species present. We report the level of genomic diversity in a natural population of Ostreococcus tauri (Chlorophyta, Mamiellophyceae), the smallest photosynthetic eukaryote. Contrary to the expectations of clonal evolution or cryptic species, the spectrum of genomic polymorphism observed suggests a large panmictic population (an effective population size of 1.2 × 107) with pervasive evidence of sexual reproduction. De novo assemblies of low-coverage chromosomes reveal two large candidate mating-type loci with suppressed recombination, whose origin may pre-date the speciation events in the class Mamiellophyceae. This high genetic diversity is associated with large phenotypic differences between strains. Strikingly, resistance of isolates to large double-stranded DNA viruses, which abound in their natural environment, is positively correlated with the size of a single hypervariable chromosome, which contains 44 to 156 kb of strain-specific sequences. Our findings highlight the role of viruses in shaping genome diversity in marine picoeukaryotes.
Keywords: GC content evolution; chromothripsis; evolutionary genomics; linkage disequilbrium; mating type locus; multiple nucleotide mutation events; picophytoplankton; population genomics; prasinovirus; sex evolution.
Figures

References
-
- Courties C., Vaquer A., Troussellier M., Lautier J., Chrétiennot-Dinet M. J., Neveux J., Machado C., Claustre H., Smallest eukaryotic organism. Nature 370, 255 (1994).
-
- Moon-van der Staay S. Y., De Wachter R., Vaulot D., Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409, 607–610 (2001). - PubMed
-
- de Vargas C., Audic S., Henry N., Decelle J., Mahé F., Logares R., Lara E., Berney C., Le Bescot N., Probert I., Carmichael M., Poulain J., Romac S., Colin S., Aury J.-M., Bittner L., Chaffron S., Dunthorn M., Engelen S., Flegontova O., Guidi L., Horák A., Jaillon O., Lima-Mendez G., Lukeš J., Malviya S., Morard R., Mulot M., Scalco E., Siano R., Vincent F., Zingone A., Dimier C., Picheral M., Searson S., Kandels-Lewis S.; Oceans Coordinators T., Acinas S. G., Bork P., Bowler C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Not F., Ogata H., Pesant S., Raes J., Sieracki M. E., Speich S., Stemmann L., Sunagawa S., Weissenbach J., Wincker P., Karsenti E., Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015). - PubMed
-
- Worden A. Z., Nolan J. K., Palenik B., Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol. Oceanogr. 49, 168–179 (2004).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

