Identifying and Validating Tankyrase Binders and Substrates: A Candidate Approach
- PMID: 28695526
- PMCID: PMC6082341
- DOI: 10.1007/978-1-4939-6993-7_28
Identifying and Validating Tankyrase Binders and Substrates: A Candidate Approach
Abstract
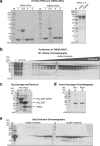
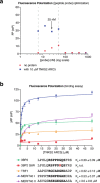
The poly(ADP-ribose)polymerase (PARP) enzyme tankyrase (TNKS/ARTD5, TNKS2/ARTD6) uses its ankyrin repeat clusters (ARCs) to recognize degenerate peptide motifs in a wide range of proteins, thereby recruiting such proteins and their complexes for scaffolding and/or poly(ADP-ribosyl)ation. Here, we provide guidance for predicting putative tankyrase-binding motifs, based on the previously delineated peptide sequence rules and existing structural information. We present a general method for the expression and purification of tankyrase ARCs from Escherichia coli and outline a fluorescence polarization assay to quantitatively assess direct ARC-TBM peptide interactions. We provide a basic protocol for evaluating binding and poly(ADP-ribosyl)ation of full-length candidate interacting proteins by full-length tankyrase in mammalian cells.
Keywords: Enzyme–substrate relationships; Fluorescence polarization (FP); PARP; Poly(ADP-ribosyl)ation; Protein expression; Protein purification; Protein-protein interactions; Structural biology; Tankyrase; Tankyrase-binding peptide motif.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

