Computational and Experimental Studies of ADP-Ribosylation
- PMID: 28695527
- PMCID: PMC5526223
- DOI: 10.1007/978-1-4939-6993-7_29
Computational and Experimental Studies of ADP-Ribosylation
Abstract
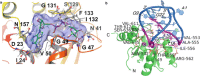
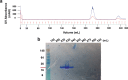
The macrodomains are a multifunctional protein family that function as receptors and enzymes acting on poly(ADP-ribose), ADP-ribosylated proteins, and other metabolites of nicotinamide adenine dinucleotide (NAD+). Several new functions for macrodomains, such as nucleic acid binding and protein-protein interaction, have recently been identified in this family. Here, we discuss methods for the identification of new macrodomains in viruses and the prediction of their function. This is followed by the expression and purification of these proteins following overexpression in bacterial cells and confirmation of folding and function using biophysical methods.
Keywords: Bioinformatics; Circular dichroism; Coronavirus; Docking; Expression; Macrodomain; NMR spectroscopy; Purification; STD-NMR.
Figures

References
-
- de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

