Relative abundance of 'Candidatus Tenderia electrophaga' is linked to cathodic current in an aerobic biocathode community
- PMID: 28696003
- PMCID: PMC5743799
- DOI: 10.1111/1751-7915.12757
Relative abundance of 'Candidatus Tenderia electrophaga' is linked to cathodic current in an aerobic biocathode community
Abstract
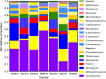
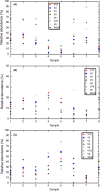
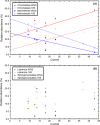
Biocathode microbial communities are proposed to catalyse a range of useful reactions. Unlike bioanodes, model biocathode organisms have not yet been successfully cultivated in isolation highlighting the need for culture-independent approaches to characterization. Biocathode MCL (Marinobacter, Chromatiaceae, Labrenzia) is a microbial community proposed to couple CO2 fixation to extracellular electron transfer and O2 reduction. Previous metagenomic analysis of a single MCL bioelectrochemical system (BES) resulted in resolution of 16 bin genomes. To further resolve bin genomes and compare community composition across replicate MCL BES, we performed shotgun metagenomic and 16S rRNA gene (16S) sequencing at steady-state current. Clustering pooled reads from replicate BES increased the number of resolved bin genomes to 20, over half of which were > 90% complete. Direct comparison of unassembled metagenomic reads and 16S operational taxonomic units (OTUs) predicted higher community diversity than the assembled/clustered metagenome and the predicted relative abundances did not match. However, when 16S OTUs were mapped to bin genomes and genome abundance was scaled by 16S gene copy number, estimated relative abundance was more similar to metagenomic analysis. The relative abundance of the bin genome representing 'Ca. Tenderia electrophaga' was correlated with increasing current, further supporting the hypothesis that this organism is the electroautotroph.
Published 2017. This article is a U.S. Government work and is in the public domain in the USA. Microbial Biotechnology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures
References
-
- Albertsen, M. , Hugenholtz, P. , Skarshewski, A. , Nielsen, K.L. , Tyson, G.W. , and Nielsen, P.H. (2013) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. - PubMed
-
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. , and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Bushnell, B. (2016) BBMap short read aligner. URL http://sourceforgenet/projects/bbmap.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

