Role and Therapeutic Targeting of the HGF/MET Pathway in Glioblastoma
- PMID: 28696366
- PMCID: PMC5532623
- DOI: 10.3390/cancers9070087
Role and Therapeutic Targeting of the HGF/MET Pathway in Glioblastoma
Abstract
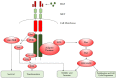
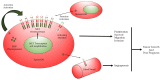
Glioblastoma (GBM) is a lethal brain tumor with dismal prognosis. Current therapeutic options, consisting of surgery, chemotherapy and radiation, have only served to marginally increase patient survival. Receptor tyrosine kinases (RTKs) are dysregulated in approximately 90% of GBM; attributed to this, research has focused on inhibiting RTKs as a novel and effective therapy for GBM. Overexpression of RTK mesenchymal epithelial transition (MET), and its ligand, hepatocyte growth factor (HGF), in GBM highlights a promising new therapeutic target. This review will discuss the role of MET in cell cycle regulation, cell proliferation, evasion of apoptosis, cell migration and invasion, angiogenesis and therapeutic resistance in GBM. It will also discuss the modes of deregulation of HGF/MET and their regulation by microRNAs. As the HGF/MET pathway is a vital regulator of multiple pro-survival pathways, efforts and strategies for its exploitation for GBM therapy are also described.
Keywords: Glioblastoma; HGF; MET.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, AND NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. - DOI - PMC - PubMed
-
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

