Functional Profiling of Transcription Factor Genes in Neurospora crassa
- PMID: 28696922
- PMCID: PMC5592922
- DOI: 10.1534/g3.117.043331
Functional Profiling of Transcription Factor Genes in Neurospora crassa
Abstract
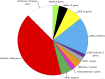
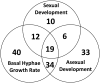
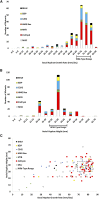
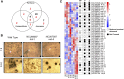
Regulation of gene expression by DNA-binding transcription factors is essential for proper control of growth and development in all organisms. In this study, we annotate and characterize growth and developmental phenotypes for transcription factor genes in the model filamentous fungus Neurospora crassa We identified 312 transcription factor genes, corresponding to 3.2% of the protein coding genes in the genome. The largest class was the fungal-specific Zn2Cys6 (C6) binuclear cluster, with 135 members, followed by the highly conserved C2H2 zinc finger group, with 61 genes. Viable knockout mutants were produced for 273 genes, and complete growth and developmental phenotypic data are available for 242 strains, with 64% possessing at least one defect. The most prominent defect observed was in growth of basal hyphae (43% of mutants analyzed), followed by asexual sporulation (38%), and the various stages of sexual development (19%). Two growth or developmental defects were observed for 21% of the mutants, while 8% were defective in all three major phenotypes tested. Analysis of available mRNA expression data for a time course of sexual development revealed mutants with sexual phenotypes that correlate with transcription factor transcript abundance in wild type. Inspection of this data also implicated cryptic roles in sexual development for several cotranscribed transcription factor genes that do not produce a phenotype when mutated.
Keywords: filamentous fungi; functional genomics; gene knockouts; transcription factors; transcriptional profiling.
Copyright © 2017 Carrillo et al.
Figures
References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Aravind L., Anantharaman V., Balaji S., Babu M. M., Iyer L. M., 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29: 231–262. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
