Whole Genome Sequence of the Heterozygous Clinical Isolate Candida krusei 81-B-5
- PMID: 28696923
- PMCID: PMC5592916
- DOI: 10.1534/g3.117.043547
Whole Genome Sequence of the Heterozygous Clinical Isolate Candida krusei 81-B-5
Abstract
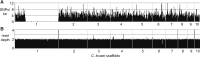
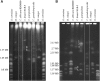
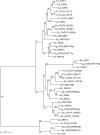
Candida krusei is a diploid, heterozygous yeast that is an opportunistic fungal pathogen in immunocompromised patients. This species also is utilized for fermenting cocoa beans during chocolate production. One major concern in the clinical setting is the innate resistance of this species to the most commonly used antifungal drug fluconazole. Here, we report a high-quality genome sequence and assembly for the first clinical isolate of C. krusei, strain 81-B-5, into 11 scaffolds generated with PacBio sequencing technology. Gene annotation and comparative analysis revealed a unique profile of transporters that could play a role in drug resistance or adaptation to different environments. In addition, we show that, while 82% of the genome is highly heterozygous, a 2.0 Mb region of the largest scaffold has undergone loss of heterozygosity. This genome will serve as a reference for further genetic studies of this pathogen.
Keywords: 81-B-5; Candida krusei; Genome Report; LOH; heterozygosity; mating type locus; transporters.
Copyright © 2017 Cuomo et al.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
