Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment
- PMID: 28697342
- PMCID: PMC5599156
- DOI: 10.1016/j.ccell.2017.06.003
Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment
Erratum in
-
Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment.Cancer Cell. 2018 Jan 8;33(1):152. doi: 10.1016/j.ccell.2017.12.012. Cancer Cell. 2018. PMID: 29316430 Free PMC article. No abstract available.
Abstract
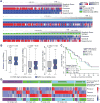
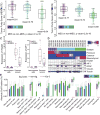
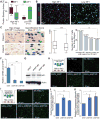
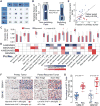
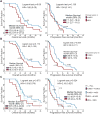
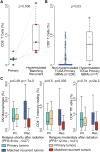
We leveraged IDH wild-type glioblastomas, derivative neurospheres, and single-cell gene expression profiles to define three tumor-intrinsic transcriptional subtypes designated as proneural, mesenchymal, and classical. Transcriptomic subtype multiplicity correlated with increased intratumoral heterogeneity and presence of tumor microenvironment. In silico cell sorting identified macrophages/microglia, CD4+ T lymphocytes, and neutrophils in the glioma microenvironment. NF1 deficiency resulted in increased tumor-associated macrophages/microglia infiltration. Longitudinal transcriptome analysis showed that expression subtype is retained in 55% of cases. Gene signature-based tumor microenvironment inference revealed a decrease in invading monocytes and a subtype-dependent increase in macrophages/microglia cells upon disease recurrence. Hypermutation at diagnosis or at recurrence associated with CD8+ T cell enrichment. Frequency of M2 macrophages detection associated with short-term relapse after radiation therapy.
Keywords: disease recurrence; glioblastoma; immune cells; macrophages/microglia; mesenchymal subtype; proneural to mesenchymal transition; tumor evolution; tumor microenvironment.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

References
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

