Rewriting the Genetic Code
- PMID: 28697669
- PMCID: PMC5772603
- DOI: 10.1146/annurev-micro-090816-093247
Rewriting the Genetic Code
Abstract
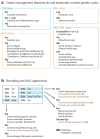
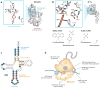
The genetic code-the language used by cells to translate their genomes into proteins that perform many cellular functions-is highly conserved throughout natural life. Rewriting the genetic code could lead to new biological functions such as expanding protein chemistries with noncanonical amino acids (ncAAs) and genetically isolating synthetic organisms from natural organisms and viruses. It has long been possible to transiently produce proteins bearing ncAAs, but stabilizing an expanded genetic code for sustained function in vivo requires an integrated approach: creating recoded genomes and introducing new translation machinery that function together without compromising viability or clashing with endogenous pathways. In this review, we discuss design considerations and technologies for expanding the genetic code. The knowledge obtained by rewriting the genetic code will deepen our understanding of how genomes are designed and how the canonical genetic code evolved.
Keywords: codon usage; genetic code; orthogonal; synthetic biology; translation engineering.
Conflict of interest statement
M.J.L. has a financial interest in GRO Biosciences and is coinventor on patents related to genomically recoded organisms.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

