A proteomic analysis of LRRK2 binding partners reveals interactions with multiple signaling components of the WNT/PCP pathway
- PMID: 28697798
- PMCID: PMC5505151
- DOI: 10.1186/s13024-017-0193-9
A proteomic analysis of LRRK2 binding partners reveals interactions with multiple signaling components of the WNT/PCP pathway
Abstract
Background: Autosomal-dominant mutations in the Park8 gene encoding Leucine-rich repeat kinase 2 (LRRK2) have been identified to cause up to 40% of the genetic forms of Parkinson's disease. However, the function and molecular pathways regulated by LRRK2 are largely unknown. It has been shown that LRRK2 serves as a scaffold during activation of WNT/β-catenin signaling via its interaction with the β-catenin destruction complex, DVL1-3 and LRP6. In this study, we examine whether LRRK2 also interacts with signaling components of the WNT/Planar Cell Polarity (WNT/PCP) pathway, which controls the maturation of substantia nigra dopaminergic neurons, the main cell type lost in Parkinson's disease patients.
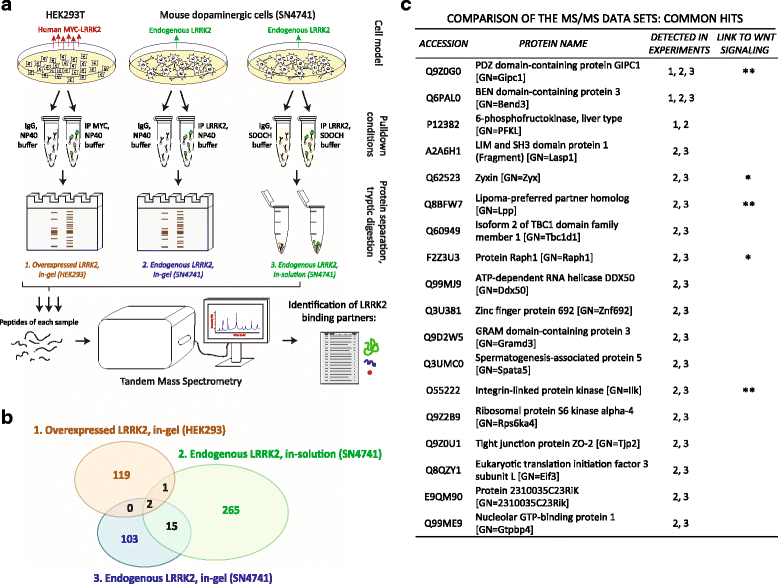
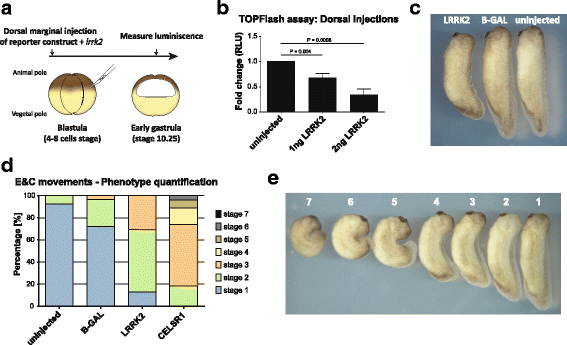
Methods: Co-immunoprecipitation and tandem mass spectrometry was performed in a mouse substantia nigra cell line (SN4741) and human HEK293T cell line in order to identify novel LRRK2 binding partners. Inhibition of the WNT/β-catenin reporter, TOPFlash, was used as a read-out of WNT/PCP pathway activation. The capacity of LRRK2 to regulate WNT/PCP signaling in vivo was tested in Xenopus laevis' early development.
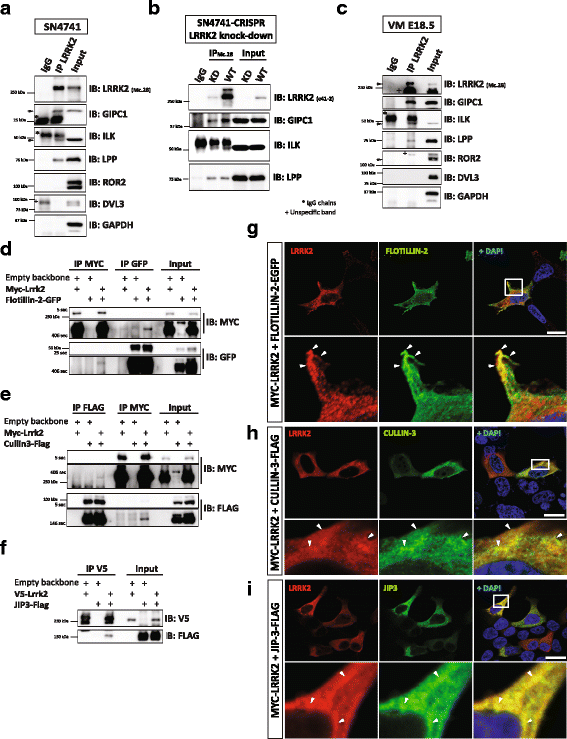
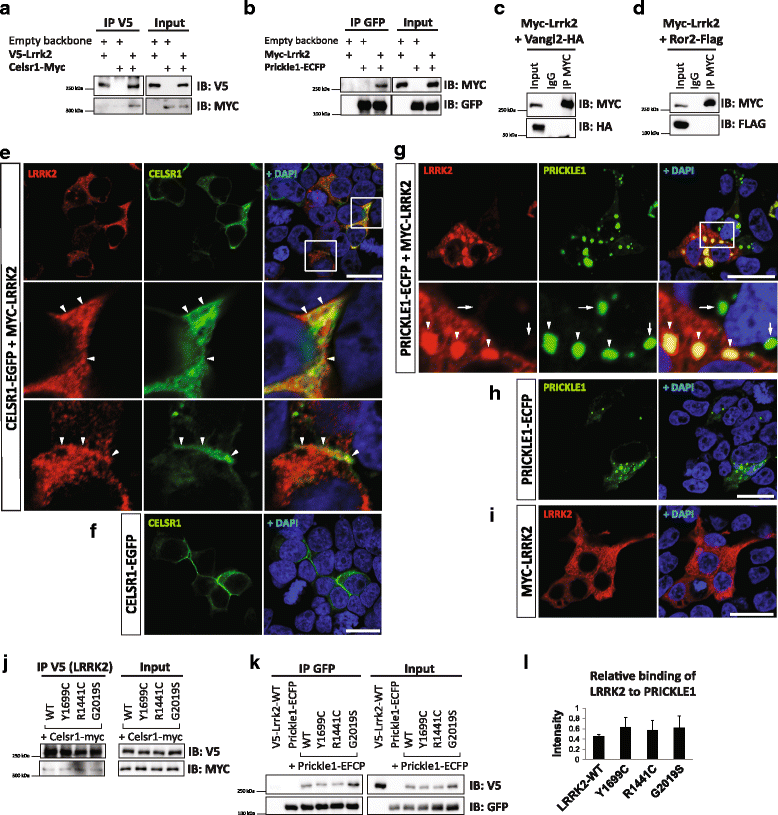
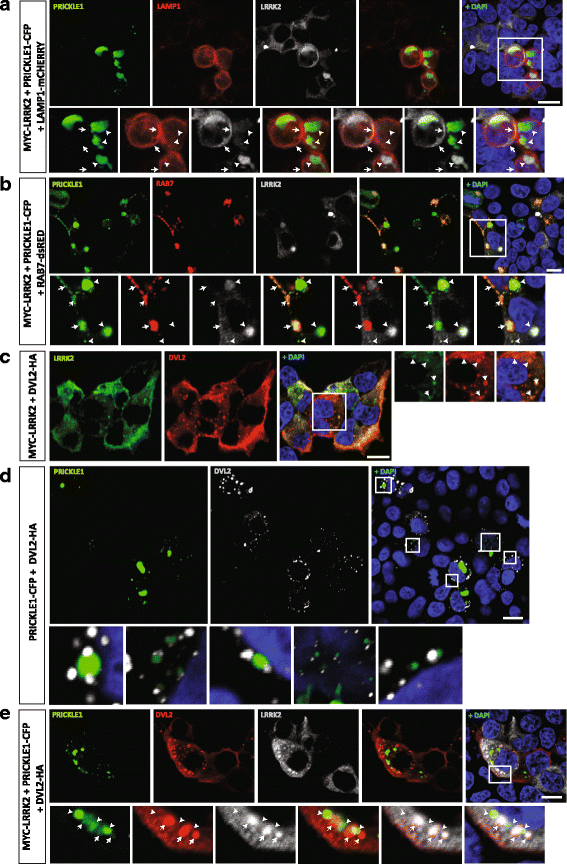
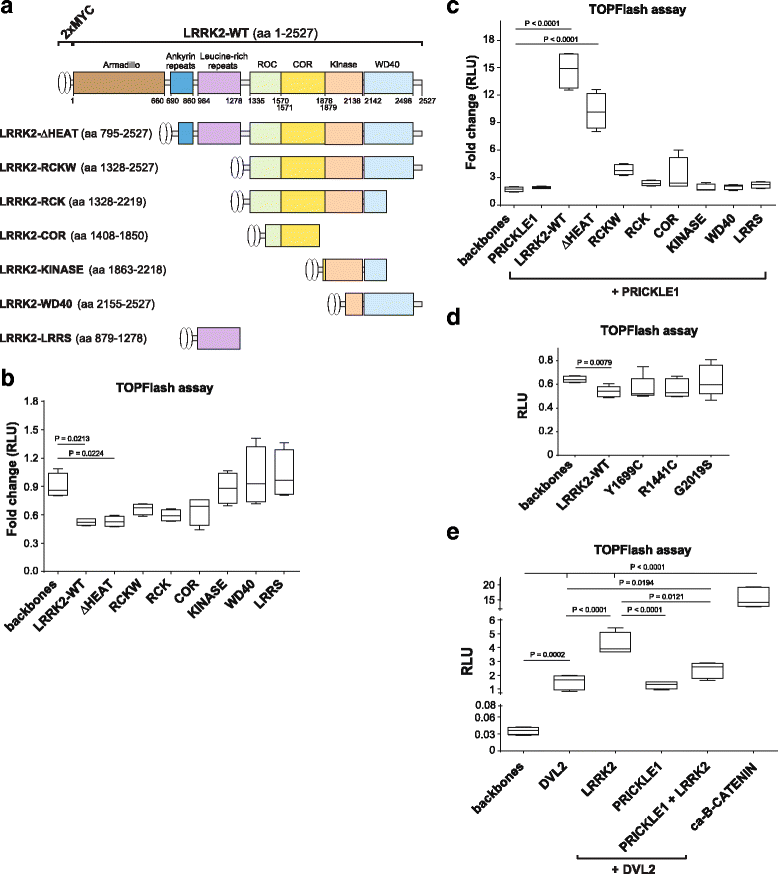
Results: Our proteomic analysis identified that LRRK2 interacts with proteins involved in WNT/PCP signaling such as the PDZ domain-containing protein GIPC1 and Integrin-linked kinase (ILK) in dopaminergic cells in vitro and in the mouse ventral midbrain in vivo. Moreover, co-immunoprecipitation analysis revealed that LRRK2 binds to two core components of the WNT/PCP signaling pathway, PRICKLE1 and CELSR1, as well as to FLOTILLIN-2 and CULLIN-3, which regulate WNT secretion and inhibit WNT/β-catenin signaling, respectively. We also found that PRICKLE1 and LRRK2 localize in signalosomes and act as dual regulators of WNT/PCP and β-catenin signaling. Accordingly, analysis of the function of LRRK2 in vivo, in X. laevis revelaed that LRKK2 not only inhibits WNT/β-catenin pathway, but induces a classical WNT/PCP phenotype in vivo.
Conclusions: Our study shows for the first time that LRRK2 activates the WNT/PCP signaling pathway through its interaction to multiple WNT/PCP components. We suggest that LRRK2 regulates the balance between WNT/β-catenin and WNT/PCP signaling, depending on the binding partners. Since this balance is crucial for homeostasis of midbrain dopaminergic neurons, we hypothesize that its alteration may contribute to the pathophysiology of Parkinson's disease.
Keywords: CELSR1; DVL; Dopaminergic neurons; Endocytosis; Immunoprecipitation; PRICKLE1; Parkinson’s disease; Signalosomes; Substantia nigra; WNT/planar cell polarity.
Conflict of interest statement
Ethics approval
Mice were housed, bred and treated according to the guidelines of the European Communities Council (directive 86/609/EEC) and the local ethics committees (Stockholm’s Norra Djurförsöketiska Nämnd N158/15).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Dachsel JC, Nishioka K, Vilarino-Guell C, Lincoln SJ, Soto-Ortolaza AI, Kachergus J, Hinkle KM, Heckman MG, Jasinska-Myga B, Taylor JP, et al. Heterodimerization of Lrrk1-Lrrk2: implications for LRRK2-associated Parkinson disease. Mech Ageing Dev. 2010;131:210–214. doi: 10.1016/j.mad.2010.01.009. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

