Dynamic graph metrics: Tutorial, toolbox, and tale
- PMID: 28698107
- PMCID: PMC5758445
- DOI: 10.1016/j.neuroimage.2017.06.081
Dynamic graph metrics: Tutorial, toolbox, and tale
Abstract
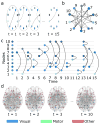
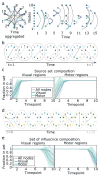
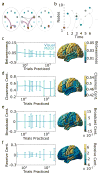
The central nervous system is composed of many individual units - from cells to areas - that are connected with one another in a complex pattern of functional interactions that supports perception, action, and cognition. One natural and parsimonious representation of such a system is a graph in which nodes (units) are connected by edges (interactions). While applicable across spatiotemporal scales, species, and cohorts, the traditional graph approach is unable to address the complexity of time-varying connectivity patterns that may be critically important for an understanding of emotional and cognitive state, task-switching, adaptation and development, or aging and disease progression. Here we survey a set of tools from applied mathematics that offer measures to characterize dynamic graphs. Along with this survey, we offer suggestions for visualization and a publicly-available MATLAB toolbox to facilitate the application of these metrics to existing or yet-to-be acquired neuroimaging data. We illustrate the toolbox by applying it to a previously published data set of time-varying functional graphs, but note that the tools can also be applied to time-varying structural graphs or to other sorts of relational data entirely. Our aim is to provide the neuroimaging community with a useful set of tools, and an intuition regarding how to use them, for addressing emerging questions that hinge on accurate and creative analyses of dynamic graphs.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
GraphVar: a user-friendly toolbox for comprehensive graph analyses of functional brain connectivity.J Neurosci Methods. 2015 Apr 30;245:107-15. doi: 10.1016/j.jneumeth.2015.02.021. Epub 2015 Feb 26. J Neurosci Methods. 2015. PMID: 25725332
-
SPectral graph theory And Random walK (SPARK) toolbox for static and dynamic characterization of (di)graphs: A tutorial.PLoS One. 2025 Jun 5;20(6):e0319031. doi: 10.1371/journal.pone.0319031. eCollection 2025. PLoS One. 2025. PMID: 40472336 Free PMC article.
-
GraphVar 2.0: A user-friendly toolbox for machine learning on functional connectivity measures.J Neurosci Methods. 2018 Oct 1;308:21-33. doi: 10.1016/j.jneumeth.2018.07.001. Epub 2018 Jul 17. J Neurosci Methods. 2018. PMID: 30026069
-
Mapping the functional connectome in traumatic brain injury: What can graph metrics tell us?Neuroimage. 2017 Oct 15;160:113-123. doi: 10.1016/j.neuroimage.2016.12.003. Epub 2016 Dec 3. Neuroimage. 2017. PMID: 27919750 Review.
-
Graph analysis of the human connectome: promise, progress, and pitfalls.Neuroimage. 2013 Oct 15;80:426-44. doi: 10.1016/j.neuroimage.2013.04.087. Epub 2013 Apr 30. Neuroimage. 2013. PMID: 23643999 Review.
Cited by
-
Brain Connectivity Studies on Structure-Function Relationships: A Short Survey with an Emphasis on Machine Learning.Comput Intell Neurosci. 2021 May 27;2021:5573740. doi: 10.1155/2021/5573740. eCollection 2021. Comput Intell Neurosci. 2021. PMID: 34135951 Free PMC article. Review.
-
On the Importance of Being Flexible: Dynamic Brain Networks and Their Potential Functional Significances.Front Syst Neurosci. 2022 Jan 21;15:688424. doi: 10.3389/fnsys.2021.688424. eCollection 2021. Front Syst Neurosci. 2022. PMID: 35126062 Free PMC article. Review.
-
Mindful attention promotes control of brain network dynamics for self-regulation and discontinues the past from the present.Proc Natl Acad Sci U S A. 2023 Jan 10;120(2):e2201074119. doi: 10.1073/pnas.2201074119. Epub 2023 Jan 3. Proc Natl Acad Sci U S A. 2023. PMID: 36595675 Free PMC article. Clinical Trial.
-
Altered resting-state dynamic functional brain networks in major depressive disorder: Findings from the REST-meta-MDD consortium.Neuroimage Clin. 2020;26:102163. doi: 10.1016/j.nicl.2020.102163. Epub 2020 Jan 7. Neuroimage Clin. 2020. PMID: 31953148 Free PMC article.
-
On the nature and use of models in network neuroscience.Nat Rev Neurosci. 2018 Sep;19(9):566-578. doi: 10.1038/s41583-018-0038-8. Nat Rev Neurosci. 2018. PMID: 30002509 Free PMC article. Review.
References
-
- Breakspear M. Dynamic models of large-scale brain activity. Nat Neurosci. 2017;20:340–352. - PubMed
-
- Villoslada P, Steinman L, Baranzini SE. Systems biology and its application to the understanding of neurological diseases. Ann Neurol. 2009;65:124–139. - PubMed
-
- Cooper RP, Shallice T. Cognitive neuroscience: the troubled marriage of cognitive science and neuroscience. Top Cogn Sci. 2010;2:398–406. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

