Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling
- PMID: 28698226
- PMCID: PMC5592811
- DOI: 10.1242/dev.147629
Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling
Abstract
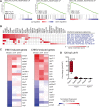
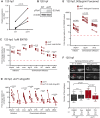
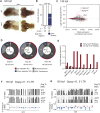
Complex cytoplasmic nucleotide-sensing mechanisms can recognize foreign DNA based on a lack of methylation and initiate an immune response to clear the infection. Zebrafish embryos with global DNA hypomethylation caused by mutations in the ubiquitin-like with PHD and ring finger domains 1 (uhrf1) or DNA methyltransferase 1 (dnmt1) genes exhibit a robust interferon induction characteristic of the first line of defense against viral infection. We found that this interferon induction occurred in non-immune cells and examined whether intracellular viral sensing pathways in these cells were the trigger. RNA-seq analysis of uhrf1 and dnmt1 mutants revealed widespread induction of Class I retrotransposons and activation of cytoplasmic DNA viral sensors. Attenuating Sting, phosphorylated Tbk1 and, importantly, blocking reverse transcriptase activity suppressed the expression of interferon genes in uhrf1 mutants. Thus, activation of transposons in cells with global DNA hypomethylation mimics a viral infection by activating cytoplasmic DNA sensors. This suggests that antiviral pathways serve as surveillance of cells that have derepressed intragenomic parasites due to DNA hypomethylation.
Keywords: Antiviral; DNA methylation; Interferon; Transposon; Uhrf1.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures





References
-
- Absher D. M., Li X., Waite L. L., Gibson A., Roberts K., Edberg J., Chatham W. W. and Kimberly R. P. (2013). Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 9, e1003678 10.1371/journal.pgen.1003678 - DOI - PMC - PubMed
-
- Aggad D., Mazel M., Boudinot P., Mogensen K. E., Hamming O. J., Hartmann R., Kotenko S., Herbomel P., Lutfalla G. and Levraud J.-P. (2009). The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J. Immunol. 183, 3924-3931. 10.4049/jimmunol.0901495 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

