Enzymatic production of single-molecule FISH and RNA capture probes
- PMID: 28698239
- PMCID: PMC5602115
- DOI: 10.1261/rna.061184.117
Enzymatic production of single-molecule FISH and RNA capture probes
Abstract
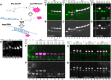
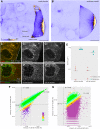
Arrays of singly labeled short oligonucleotides that hybridize to a specific target revolutionized RNA biology, enabling quantitative, single-molecule microscopy analysis and high-efficiency RNA/RNP capture. Here, we describe a simple and efficient method that allows flexible functionalization of inexpensive DNA oligonucleotides by different fluorescent dyes or biotin using terminal deoxynucleotidyl transferase and custom-made functional group conjugated dideoxy-UTP. We show that (i) all steps of the oligonucleotide labeling-including conjugation, enzymatic synthesis, and product purification-can be performed in a standard biology laboratory, (ii) the process yields >90%, often >95% labeled product with minimal carryover of impurities, and (iii) the oligonucleotides can be labeled with different dyes or biotin, allowing single-molecule FISH, RNA affinity purification, and Northern blot analysis to be performed.
Keywords: RNA affinity purification; dideoxy-UTP; oligonucleotide labeling; single-molecule FISH; terminal deoxynucleotidyl transferase.
© 2017 Gaspar et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Femino AM, Fay FS, Fogarty K, Singer RH. 1998. Visualization of single RNA transcripts in situ. Science 280: 585–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
