Neat1 is a p53-inducible lincRNA essential for transformation suppression
- PMID: 28698299
- PMCID: PMC5538433
- DOI: 10.1101/gad.284661.116
Neat1 is a p53-inducible lincRNA essential for transformation suppression
Abstract
The p53 gene is mutated in over half of all cancers, reflecting its critical role as a tumor suppressor. Although p53 is a transcriptional activator that induces myriad target genes, those p53-inducible genes most critical for tumor suppression remain elusive. Here, we leveraged p53 ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) and RNA-seq (RNA sequencing) data sets to identify new p53 target genes, focusing on the noncoding genome. We identify Neat1, a noncoding RNA (ncRNA) constituent of paraspeckles, as a p53 target gene broadly induced by mouse and human p53 in different cell types and by diverse stress signals. Using fibroblasts derived from Neat1-/- mice, we examined the functional role of Neat1 in the p53 pathway. We found that Neat1 is dispensable for cell cycle arrest and apoptosis in response to genotoxic stress. In sharp contrast, Neat1 plays a crucial role in suppressing transformation in response to oncogenic signals. Neat1 deficiency enhances transformation in oncogene-expressing fibroblasts and promotes the development of premalignant pancreatic intraepithelial neoplasias (PanINs) and cystic lesions in KrasG12D-expressing mice. Neat1 loss provokes global changes in gene expression, suggesting a mechanism by which its deficiency promotes neoplasia. Collectively, these findings identify Neat1 as a p53-regulated large intergenic ncRNA (lincRNA) with a key role in suppressing transformation and cancer initiation, providing fundamental new insight into p53-mediated tumor suppression.
Keywords: Neat1; lincRNA; p53; pancreatic cancer; tumor suppression.
© 2017 Mello et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

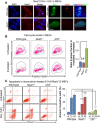
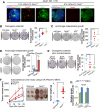
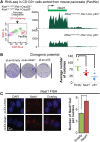
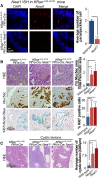
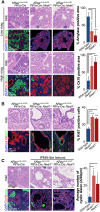
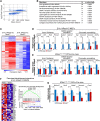
Comment in
-
Cancer biology: A Neat target of p53.Nat Rev Mol Cell Biol. 2017 Sep;18(9):532. doi: 10.1038/nrm.2017.80. Epub 2017 Aug 2. Nat Rev Mol Cell Biol. 2017. PMID: 28792010 No abstract available.
References
-
- Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PWG, Radaelli E, Vermi W, et al. 2016. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med 22: 861–868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
