A distinct subpopulation of CD25- T-follicular regulatory cells localizes in the germinal centers
- PMID: 28698369
- PMCID: PMC5547636
- DOI: 10.1073/pnas.1705551114
A distinct subpopulation of CD25- T-follicular regulatory cells localizes in the germinal centers
Abstract
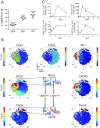
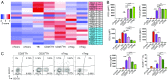
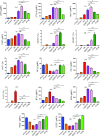
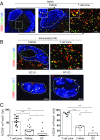
T-follicular helper (Tfh) cells differentiate through a multistep process, culminating in germinal center (GC) localized GC-Tfh cells that provide support to GC-B cells. T-follicular regulatory (Tfr) cells have critical roles in the control of Tfh cells and GC formation. Although Tfh-cell differentiation is inhibited by IL-2, regulatory T (Treg) cell differentiation and survival depend on it. Here, we describe a CD25- subpopulation within both murine and human PD1+CXCR5+Foxp3+ Tfr cells. It is preferentially located in the GC and can be clearly differentiated from CD25+ non-GC-Tfr, Tfh, and effector Treg (eTreg) cells by the expression of a wide range of molecules. In comparison to CD25+ Tfr and eTreg cells, CD25- Tfr cells partially down-regulate IL-2-dependent canonical Treg features, but retain suppressive function, while simultaneously up-regulating genes associated with Tfh and GC-Tfh cells. We suggest that, similar to Tfh cells, Tfr cells follow a differentiation pathway generating a mature GC-localized subpopulation, CD25- Tfr cells.
Keywords: T-follicular regulatory cell; Tfh cell; Tfr cell; Treg cell; germinal center.
Conflict of interest statement
Conflict of interest statement: S.H. and S.S. have independently collaborated with the same laboratory and have therefore appeared as coauthors in a paper published as Garg G, et al. (2017) Unique properties of thymic antigen-presenting cells promote epigenetic imprinting of alloantigen-specific regulatory T cells. Oncotarget 8:35542–35557, although S.S. did so independently and without knowledge of the other's participation in the work, S.H. and S.S. contributed independently to an article regarding nomenclature published as Abbas AK, et al. (2013) Regulatory T cells: Recommendations to simplify the nomenclature. Nat Immunol 14:307–308.
Figures

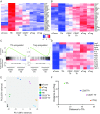
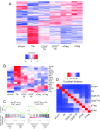


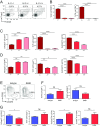
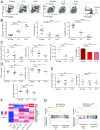


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

