Deletion of Gas2l3 in mice leads to specific defects in cardiomyocyte cytokinesis during development
- PMID: 28698371
- PMCID: PMC5544300
- DOI: 10.1073/pnas.1703406114
Deletion of Gas2l3 in mice leads to specific defects in cardiomyocyte cytokinesis during development
Abstract
GAS2L3 is a recently identified cytoskeleton-associated protein that interacts with actin filaments and tubulin. The in vivo function of GAS2L3 in mammals remains unknown. Here, we show that mice deficient in GAS2L3 die shortly after birth because of heart failure. Mammalian cardiomyocytes lose the ability to proliferate shortly after birth, and further increase in cardiac mass is achieved by hypertrophy. The proliferation arrest of cardiomyocytes is accompanied by binucleation through incomplete cytokinesis. We observed that GAS2L3 deficiency leads to inhibition of cardiomyocyte proliferation and to cardiomyocyte hypertrophy during embryonic development. Cardiomyocyte-specific deletion of GAS2L3 confirmed that the phenotype results from the loss of GAS2L3 in cardiomyocytes. Cardiomyocytes from Gas2l3-deficient mice exhibit increased expression of a p53-transcriptional program including the cell cycle inhibitor p21. Furthermore, loss of GAS2L3 results in premature binucleation of cardiomyocytes accompanied by unresolved midbody structures. Together these results suggest that GAS2L3 plays a specific role in cardiomyocyte cytokinesis and proliferation during heart development.
Keywords: GAS2L3; binucleation; cardiomyocytes; cytokinesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

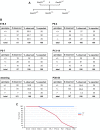



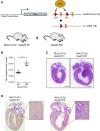
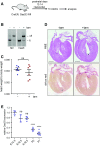
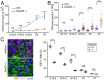

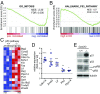
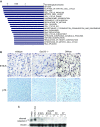
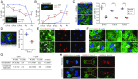
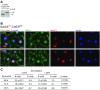
Comment in
-
GAS2L3: Coordinator of cardiomyocyte cytokinesis?Cell Cycle. 2017 Oct 18;16(20):1853-1854. doi: 10.1080/15384101.2017.1372546. Epub 2017 Sep 22. Cell Cycle. 2017. PMID: 28937871 Free PMC article. No abstract available.
References
-
- Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. - PubMed
-
- Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. - PubMed
-
- Zebrowski DC, Engel FB. The cardiomyocyte cell cycle in hypertrophy, tissue homeostasis, and regeneration. Rev Physiol Biochem Pharmacol. 2013;165:67–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

