Delayed Cryptochrome Degradation Asymmetrically Alters the Daily Rhythm in Suprachiasmatic Clock Neuron Excitability
- PMID: 28698388
- PMCID: PMC5559760
- DOI: 10.1523/JNEUROSCI.0691-17.2017
Delayed Cryptochrome Degradation Asymmetrically Alters the Daily Rhythm in Suprachiasmatic Clock Neuron Excitability
Abstract
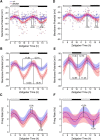
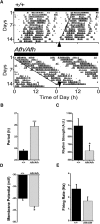
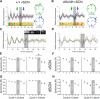
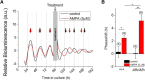
Suprachiasmatic nuclei (SCN) neurons contain an intracellular molecular circadian clock and the Cryptochromes (CRY1/2), key transcriptional repressors of this molecular apparatus, are subject to post-translational modification through ubiquitination and targeting for proteosomal degradation by the ubiquitin E3 ligase complex. Loss-of-function point mutations in a component of this ligase complex, Fbxl3, delay CRY1/2 degradation, reduce circadian rhythm strength, and lengthen the circadian period by ∼2.5 h. The molecular clock drives circadian changes in the membrane properties of SCN neurons, but it is unclear how alterations in CRY1/2 stability affect SCN neurophysiology. Here we use male and female Afterhours mice which carry the circadian period lengthening loss-of-function Fbxl3Afh mutation and perform patch-clamp recordings from SCN brain slices across the projected day/night cycle. We find that the daily rhythm in membrane excitability in the ventral SCN (vSCN) was enhanced in amplitude and delayed in timing in Fbxl3Afh/Afh mice. At night, vSCN cells from Fbxl3Afh/Afh mice were more hyperpolarized, receiving more GABAergic input than their Fbxl3+/+ counterparts. Unexpectedly, the progression to daytime hyperexcited states was slowed by Afh mutation, whereas the decline to hypoexcited states was accelerated. In long-term bioluminescence recordings, GABAA receptor blockade desynchronized the Fbxl3+/+ but not the Fbxl3Afh/Afh vSCN neuronal network. Further, a neurochemical mimic of the light input pathway evoked larger shifts in molecular clock rhythms in Fbxl3Afh/Afh compared with Fbxl3+/+ SCN slices. These results reveal unanticipated consequences of delaying CRY degradation, indicating that the Afh mutation prolongs nighttime hyperpolarized states of vSCN cells through increased GABAergic synaptic transmission.SIGNIFICANCE STATEMENT The intracellular molecular clock drives changes in SCN neuronal excitability, but it is unclear how mutations affecting post-translational modification of molecular clock proteins influence the temporal expression of SCN neuronal state or intercellular communication within the SCN network. Here we show for the first time, that a mutation that prolongs the stability of key components of the intracellular clock, the cryptochrome proteins, unexpectedly increases in the expression of hypoexcited neuronal state in the ventral SCN at night and enhances hyperpolarization of ventral SCN neurons at this time. This is accompanied by increased GABAergic signaling and by enhanced responsiveness to a neurochemical mimic of the light input pathway to the SCN. Therefore, post-translational modification shapes SCN neuronal state and network properties.
Keywords: brain slice; circadian; cryptochrome; electrophysiology; fbxl3; post-translational modification; suprachiasmatic.
Copyright © 2017 Wegner et al.
Figures


Similar articles
-
Distinct and separable roles for endogenous CRY1 and CRY2 within the circadian molecular clockwork of the suprachiasmatic nucleus, as revealed by the Fbxl3(Afh) mutation.J Neurosci. 2013 Apr 24;33(17):7145-53. doi: 10.1523/JNEUROSCI.4950-12.2013. J Neurosci. 2013. PMID: 23616524 Free PMC article.
-
Rhythmic expression of cryptochrome induces the circadian clock of arrhythmic suprachiasmatic nuclei through arginine vasopressin signaling.Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2732-7. doi: 10.1073/pnas.1519044113. Epub 2016 Feb 22. Proc Natl Acad Sci U S A. 2016. PMID: 26903624 Free PMC article.
-
Translational switching of Cry1 protein expression confers reversible control of circadian behavior in arrhythmic Cry-deficient mice.Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):E12388-E12397. doi: 10.1073/pnas.1811438115. Epub 2018 Nov 28. Proc Natl Acad Sci U S A. 2018. PMID: 30487216 Free PMC article.
-
Misalignment of Circadian Rhythms in Diet-Induced Obesity.Adv Exp Med Biol. 2024;1460:27-71. doi: 10.1007/978-3-031-63657-8_2. Adv Exp Med Biol. 2024. PMID: 39287848 Review.
-
Molecular-genetic Manipulation of the Suprachiasmatic Nucleus Circadian Clock.J Mol Biol. 2020 May 29;432(12):3639-3660. doi: 10.1016/j.jmb.2020.01.019. Epub 2020 Jan 26. J Mol Biol. 2020. PMID: 31996314 Review.
Cited by
-
Loss of neuropeptide signalling alters temporal expression of mouse suprachiasmatic neuronal state and excitability.Eur J Neurosci. 2024 Dec;60(11):6617-6633. doi: 10.1111/ejn.16590. Epub 2024 Nov 17. Eur J Neurosci. 2024. PMID: 39551976 Free PMC article.
-
Cell-cell coupling and DNA methylation abnormal phenotypes in the after-hours mice.Epigenetics Chromatin. 2021 Jan 6;14(1):1. doi: 10.1186/s13072-020-00373-5. Epigenetics Chromatin. 2021. PMID: 33407878 Free PMC article.
-
Sleep/Wake Disruption in a Mouse Model of BLOC-1 Deficiency.Front Neurosci. 2018 Nov 15;12:759. doi: 10.3389/fnins.2018.00759. eCollection 2018. Front Neurosci. 2018. PMID: 30498428 Free PMC article.
-
Timed exercise stabilizes behavioral rhythms but not molecular programs in the brain's suprachiasmatic clock.iScience. 2023 Jan 18;26(2):106002. doi: 10.1016/j.isci.2023.106002. eCollection 2023 Feb 17. iScience. 2023. PMID: 36866044 Free PMC article.
-
Contribution of membrane-associated oscillators to biological timing at different timescales.Front Physiol. 2024 Jan 9;14:1243455. doi: 10.3389/fphys.2023.1243455. eCollection 2023. Front Physiol. 2024. PMID: 38264332 Free PMC article.
References
-
- Anand SN, Maywood ES, Chesham JE, Joynson G, Banks GT, Hastings MH, Nolan PM (2013) Distinct and separable roles for endogenous CRY1 and CRY2 within the circadian molecular clockwork of the suprachiasmatic nucleus, as revealed by the Fbxl3(Afh) mutation. J Neurosci 33:7145–7153. 10.1523/JNEUROSCI.4950-12.2013 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
