Transglycosylation by a chitinase from Enterobacter cloacae subsp. cloacae generates longer chitin oligosaccharides
- PMID: 28698589
- PMCID: PMC5505975
- DOI: 10.1038/s41598-017-05140-3
Transglycosylation by a chitinase from Enterobacter cloacae subsp. cloacae generates longer chitin oligosaccharides
Abstract
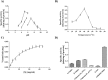
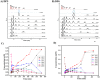
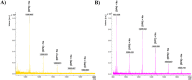
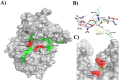
Humans have exploited natural resources for a variety of applications. Chitin and its derivative chitin oligosaccharides (CHOS) have potential biomedical and agricultural applications. Availability of CHOS with the desired length has been a major limitation in the optimum use of such natural resources. Here, we report a single domain hyper-transglycosylating chitinase, which generates longer CHOS, from Enterobacter cloacae subsp. cloacae 13047 (EcChi1). EcChi1 was optimally active at pH 5.0 and 40 °C with a Km of 15.2 mg ml-1, and k cat/Km of 0.011× 102 mg-1 ml min-1 on colloidal chitin. The profile of the hydrolytic products, major product being chitobiose, released from CHOS indicated that EcChi1 was an endo-acting enzyme. Transglycosylation (TG) by EcChi1 on trimeric to hexameric CHOS resulted in the formation of longer CHOS for a prolonged duration. EcChi1 showed both chitobiase and TG activities, in addition to hydrolytic activity. The TG by EcChi1 was dependent, to some extent, on the length of the CHOS substrate and concentration of the enzyme. Homology modeling and docking with CHOS suggested that EcChi1 has a deep substrate-binding groove lined with aromatic amino acids, which is a characteristic feature of a processive enzyme.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Pretreatment with KOH and KOH-urea enhanced hydrolysis of α-chitin by an endo-chitinase from Enterobacter cloacae subsp. cloacae.Carbohydr Polym. 2020 May 1;235:115952. doi: 10.1016/j.carbpol.2020.115952. Epub 2020 Feb 8. Carbohydr Polym. 2020. PMID: 32122487
-
Synthesis of long-chain chitooligosaccharides by a hypertransglycosylating processive endochitinase of Serratia proteamaculans 568.J Bacteriol. 2012 Aug;194(16):4260-71. doi: 10.1128/JB.06473-11. Epub 2012 Jun 8. J Bacteriol. 2012. PMID: 22685288 Free PMC article.
-
Selection and mutational analyses of the substrate interacting residues of a chitinase from Enterobacter cloacae subsp. cloacae (EcChi2) to improve transglycosylation.Int J Biol Macromol. 2020 Dec 15;165(Pt B):2432-2441. doi: 10.1016/j.ijbiomac.2020.10.125. Epub 2020 Oct 20. Int J Biol Macromol. 2020. PMID: 33096170
-
The chitinolytic machinery of Serratia marcescens--a model system for enzymatic degradation of recalcitrant polysaccharides.FEBS J. 2013 Jul;280(13):3028-49. doi: 10.1111/febs.12181. Epub 2013 Mar 7. FEBS J. 2013. PMID: 23398882 Review.
-
Production of chitooligosaccharides and their potential applications in medicine.Mar Drugs. 2010 Apr 27;8(5):1482-517. doi: 10.3390/md8051482. Mar Drugs. 2010. PMID: 20559485 Free PMC article. Review.
Cited by
-
Enzymatic Modification of Native Chitin and Conversion to Specialty Chemical Products.Mar Drugs. 2020 Jan 30;18(2):93. doi: 10.3390/md18020093. Mar Drugs. 2020. PMID: 32019265 Free PMC article. Review.
-
Bioprospecting for polyesterase activity relevant for PET degradation in marine Enterobacterales isolates.AIMS Microbiol. 2023 Jun 15;9(3):518-539. doi: 10.3934/microbiol.2023027. eCollection 2023. AIMS Microbiol. 2023. PMID: 37649797 Free PMC article.
-
Chitinase-Assisted Bioconversion of Chitinous Waste for Development of Value-Added Chito-Oligosaccharides Products.Biology (Basel). 2023 Jan 5;12(1):87. doi: 10.3390/biology12010087. Biology (Basel). 2023. PMID: 36671779 Free PMC article. Review.
-
Molecular characterization of a novel chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and its use in N-acetyl-d-glucosamine production.Biotechnol Biofuels. 2018 Jun 26;11:179. doi: 10.1186/s13068-018-1169-x. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29983742 Free PMC article.
-
Chemoenzymatic Production and Engineering of Chitooligosaccharides and N-acetyl Glucosamine for Refining Biological Activities.Front Chem. 2020 Jun 24;8:469. doi: 10.3389/fchem.2020.00469. eCollection 2020. Front Chem. 2020. PMID: 32671017 Free PMC article. Review.
References
-
- Hamed I, Ozogul F, Regenstein JM. Industrial applications of crustacean by products (chitin, chitosan, and chitooligosaccharides): A review. Trends Food Sci. Technol. 2016;48:40–50. doi: 10.1016/j.tifs.2015.11.007. - DOI
-
- Gillard L, Tran AT, Boyer FD, Beau JM. Chitooligosaccharide Synthesis Using an Ionic Tag. European J. Org. Chem. 2016;2016:1103–1109. doi: 10.1002/ejoc.201501476. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

