New technologies accelerate the exploration of non-coding RNAs in horticultural plants
- PMID: 28698797
- PMCID: PMC5496985
- DOI: 10.1038/hortres.2017.31
New technologies accelerate the exploration of non-coding RNAs in horticultural plants
Abstract
Non-coding RNAs (ncRNAs), that is, RNAs not translated into proteins, are crucial regulators of a variety of biological processes in plants. While protein-encoding genes have been relatively well-annotated in sequenced genomes, accounting for a small portion of the genome space in plants, the universe of plant ncRNAs is rapidly expanding. Recent advances in experimental and computational technologies have generated a great momentum for discovery and functional characterization of ncRNAs. Here we summarize the classification and known biological functions of plant ncRNAs, review the application of next-generation sequencing (NGS) technology and ribosome profiling technology to ncRNA discovery in horticultural plants and discuss the application of new technologies, especially the new genome-editing tool clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) systems, to functional characterization of plant ncRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
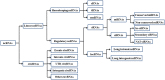
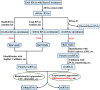
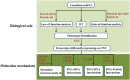
References
-
- Shin S-Y, Shin C. Regulatory non-coding RNAs in plants: potential gene resources for the improvement of agricultural traits. Plant Biotechnol Rep 2016; 10: 35–47.
-
- Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci 2012; 17: 196–203. - PubMed
-
- Holley RW, Apgar J, Everett GA et al. Structure of a ribonucleic acid. Science 1965; 147: 1462–1465. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

