Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence
- PMID: 28699239
- PMCID: PMC5595671
- DOI: 10.1111/acel.12632
Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence
Abstract
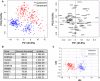
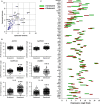
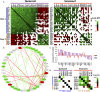
Senescent cells play important roles in both physiological and pathological processes, including cancer and aging. In all cases, however, senescent cells comprise only a small fraction of tissues. Senescent phenotypes have been studied largely in relatively homogeneous populations of cultured cells. In vivo, senescent cells are generally identified by a small number of markers, but whether and how these markers vary among individual cells is unknown. We therefore utilized a combination of single-cell isolation and a nanofluidic PCR platform to determine the contributions of individual cells to the overall gene expression profile of senescent human fibroblast populations. Individual senescent cells were surprisingly heterogeneous in their gene expression signatures. This cell-to-cell variability resulted in a loss of correlation among the expression of several senescence-associated genes. Many genes encoding senescence-associated secretory phenotype (SASP) factors, a major contributor to the effects of senescent cells in vivo, showed marked variability with a subset of highly induced genes accounting for the increases observed at the population level. Inflammatory genes in clustered genomic loci showed a greater correlation with senescence compared to nonclustered loci, suggesting that these genes are coregulated by genomic location. Together, these data offer new insights into how genes are regulated in senescent cells and suggest that single markers are inadequate to identify senescent cells in vivo.
Keywords: aging; cellular senescence; cytokines; single cell; transcriptomics.
© 2017 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures

References
-
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J (2006) Increased cell‐to‐cell variation in gene expression in ageing mouse heart. Nature 441, 1011–1014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

