Utilization of genetic data can improve the prediction of type 2 diabetes incidence in a Swedish cohort
- PMID: 28700623
- PMCID: PMC5507496
- DOI: 10.1371/journal.pone.0180180
Utilization of genetic data can improve the prediction of type 2 diabetes incidence in a Swedish cohort
Abstract
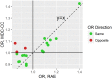
The aim of this study was to measure the impact of genetic data in improving the prediction of type 2 diabetes (T2D) in the Malmö Diet and Cancer Study cohort. The current study was performed in 3,426 Swedish individuals and utilizes of a set of genetic and environmental risk data. We first validated our environmental risk model by comparing it to both the Finnish Diabetes Risk Score and the T2D risk model derived from the Framingham Offspring Study. The area under the curve (AUC) for our environmental model was 0.72 [95% CI, 0.69-0.74], which was significantly better than both the Finnish (0.64 [95% CI, 0.61-0.66], p-value < 1 x 10-4) and Framingham (0.69 [95% CI, 0.66-0.71], p-value = 0.0017) risk scores. We then verified that the genetic data has a statistically significant positive correlation with incidence of T2D in the studied population. We also verified that adding genetic data slightly but statistically increased the AUC of a model based only on environmental risk factors (RFs, AUC shift +1.0% from 0.72 to 0.73, p-value = 0.042). To study the dependence of the results on the environmental RFs, we divided the population into two equally sized risk groups based only on their environmental risk and repeated the same analysis within each subpopulation. While there is a statistically significant positive correlation between the genetic data and incidence of T2D in both environmental risk categories, the positive shift in the AUC remains statistically significant only in the category with the lower environmental risk. These results demonstrate that genetic data can be used to increase the accuracy of T2D prediction. Also, the data suggests that genetic data is more valuable in improving T2D prediction in populations with lower environmental risk. This suggests that the impact of genetic data depends on the environmental risk of the studied population and thus genetic association studies should be performed in light of the underlying environmental risk of the population.
Conflict of interest statement
Figures



References
-
- World Health Organization: Global report on diabetes. 2016. ISBN: 978-92-4-156525-7.
-
- Murea M, Ma L, Freedman BI. Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev Diabet Stud. 2012. May 10;9(1):6–22. doi: 10.1900/RDS.2012.9.6 - DOI - PMC - PubMed
-
- Rich SS, Onengut-Gumuscu S, Concannon P. Recent progress in the genetics of diabetes. Hormone Research in Paediatrics. 2009. January 21;71(Suppl. 1):17–23. - PubMed
-
- Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Archives of internal medicine. 2007. May 28;167(10):1068–74. doi: 10.1001/archinte.167.10.1068 - DOI - PubMed
-
- Lindström J, Tuomilehto J. The diabetes risk score. Diabetes care. 2003. March 1;26(3):725–31. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

